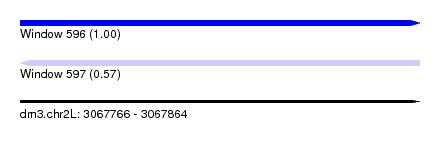
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,067,766 – 3,067,864 |
| Length | 98 |
| Max. P | 0.999593 |

| Location | 3,067,766 – 3,067,864 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 66.75 |
| Shannon entropy | 0.67004 |
| G+C content | 0.49441 |
| Mean single sequence MFE | -35.22 |
| Consensus MFE | -15.56 |
| Energy contribution | -14.31 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.45 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.06 |
| SVM RNA-class probability | 0.999593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
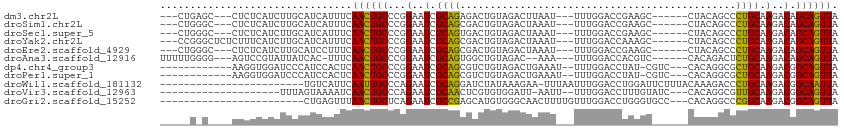
>dm3.chr2L 3067766 98 + 23011544 UAACUGCUGUCUUGCAGGGCUGUAG------GCUUCGGUCCAAA---AUUAAGUCUACAGUCUCUGCGAUUCCGGGCAGUUGAAAUGAUGCAAGAUGAGAG---GCUCAG--- (((((((((..((((((((((((((------((((.(((.....---))))))))))))))).))))))..)..))))))))...(((.((..........---))))).--- ( -36.00, z-score = -2.61, R) >droSim1.chr2L 3011071 98 + 22036055 UAACUGCUGUCUUGCAGGGCUGUAG------GCUUCGGUCCAAA---AUUUAGUCUACAGUCGCUGCGAUUCCGGGCAGUUGAAAUGAUGCAAGAUGAGAG---GCCCAG--- ...((.((((((((((.((((((((------(((..........---....)))))))))))(((((........)))))........)))))))).))))---......--- ( -33.74, z-score = -1.51, R) >droSec1.super_5 1225009 98 + 5866729 UAACUGCUGUCUUGCAGGGCUGUAG------GCUUCGGUCCAAA---AUUUAGUCUACAGUCACUGCGAUUCCGGGCAGUUGAAAUGAUGCAAGAUGAGAG---GCCCAG--- (((((((((..((((((((((((((------(((..........---....))))))))))).))))))..)..))))))))...................---......--- ( -33.64, z-score = -1.78, R) >droYak2.chr2L 3063753 101 + 22324452 UAACUGCUGUCUUGCAGGGCUGUAG------GCUUUGGUCCAAA---AUUUAGUCUACAGUCGCUGCGAUUCCGGGCAGUUGAAAUGAUGCAAGAUGAAAGAGAGCCCGG--- ...........((((((((((((((------(((..(.......---.)..))))))))))).))))))..((((((..((...((........))....))..))))))--- ( -34.20, z-score = -1.70, R) >droEre2.scaffold_4929 3115497 98 + 26641161 UAACUGCUGUCUUGCAGGGCUGUAG------GCUUCGGUCCAAA---AUUUAGUCUACAGUCGCUGCGAUUCCGGGCAGUUGAAAGGAUGCAAGAUGAGAG---GCCCAG--- (((((((((..((((((((((((((------(((..........---....))))))))))).))))))..)..))))))))...((..((..........---))))..--- ( -34.34, z-score = -1.41, R) >droAna3.scaffold_12916 6592632 98 - 16180835 UAACUGCUGUCUUGCAGAGUCUGUG------GACGUGGUCCAAA---UUU--GUCUACAGCCACUGCGAUUCCGGGCAGUUGAAA-GUGAUAAUACGGACU---CCCCAAAAA (((((((((..((((((.(.(((((------((((.........---..)--)))))))).).))))))..)..))))))))...-..........((...---..))..... ( -30.70, z-score = -2.08, R) >dp4.chr4_group3 4526508 95 - 11692001 UAACUGCCGUCUUGCAGCGCCUGUG---GACG-AUAGGUCCAAA--AUUUCAGUCUACAGACGCUGCGAUUCCGGGCAGUUGAGUGGAUGGGAUCCACCUU------------ (((((((((..((((((((.(((((---((((-(..........--...)).)))))))).))))))))..)..)))))))).((((((...))))))...------------ ( -42.72, z-score = -4.37, R) >droPer1.super_1 1669963 95 - 10282868 UAACUGCCGUCUUGCAGCGCCUGUG---GACG-AUAGGUCCAAA--AUUUCAGUCUACAGACGCUGCGAUUCCGGGCAGUUGAGUGGAUGGGAUCCACCUU------------ (((((((((..((((((((.(((((---((((-(..........--...)).)))))))).))))))))..)..)))))))).((((((...))))))...------------ ( -42.72, z-score = -4.37, R) >droWil1.scaffold_181132 407342 88 + 1035393 UAAUUGCCGUCUUGCAGGGUCUUUGUAAAGAAUCCAGGUCCAAAUUAAA-UUCUUUAUAGAUCCUGCGAUUCUGGGCAAUUGAAUGACA------------------------ (((((((((..(((((((((((..(((((((((..((.......))..)-)))))))))))))))))))..)..)))))))).......------------------------ ( -30.90, z-score = -4.53, R) >droVir3.scaffold_12963 5738683 87 - 20206255 UAACUGCCGUCUUGCAACGCCUGUG---GAUACAAAGGUCCAAA--AAUU-AAUCCACACGAGUUGCGAUUCUGGGCAGUUGAUUUUACUAAA-------------------- (((((((((..(((((((.(.((((---(((.............--....-.))))))).).)))))))..)..))))))))...........-------------------- ( -28.57, z-score = -3.76, R) >droGri2.scaffold_15252 2029281 86 + 17193109 UAACUGCCGUCUUGCCGGGCCUGUG---GGCACCCAGGUCCAAACAAAAGUUGCCCACAUGCUCGGCGAUUCUGAGCAGUUAAACUCAG------------------------ (((((((((..(((((((((.((((---((((.(...((....))....).)))))))).)))))))))..).).))))))).......------------------------ ( -39.90, z-score = -5.05, R) >consensus UAACUGCUGUCUUGCAGGGCCUUUG______GCUUAGGUCCAAA___AUUUAGUCUACAGUCGCUGCGAUUCCGGGCAGUUGAAAUGAUGCAAGAUGAGAG____C_C_____ (((((((((..((((((.((((.............)))).............(....).....))))))..)..))))))))............................... (-15.56 = -14.31 + -1.25)

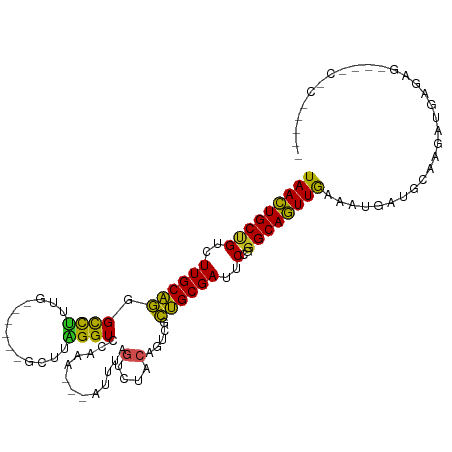
| Location | 3,067,766 – 3,067,864 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 66.75 |
| Shannon entropy | 0.67004 |
| G+C content | 0.49441 |
| Mean single sequence MFE | -28.01 |
| Consensus MFE | -8.68 |
| Energy contribution | -8.19 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.571807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3067766 98 - 23011544 ---CUGAGC---CUCUCAUCUUGCAUCAUUUCAACUGCCCGGAAUCGCAGAGACUGUAGACUUAAU---UUUGGACCGAAGC------CUACAGCCCUGCAAGACAGCAGUUA ---.((((.---..))))..............((((((..(.....((((.(.((((((.(((...---.........))).------))))))).))))....).)))))). ( -22.00, z-score = -0.15, R) >droSim1.chr2L 3011071 98 - 22036055 ---CUGGGC---CUCUCAUCUUGCAUCAUUUCAACUGCCCGGAAUCGCAGCGACUGUAGACUAAAU---UUUGGACCGAAGC------CUACAGCCCUGCAAGACAGCAGUUA ---((((((---........................))))))....((((.(.((((((.((....---..........)).------)))))).)))))............. ( -21.80, z-score = 0.60, R) >droSec1.super_5 1225009 98 - 5866729 ---CUGGGC---CUCUCAUCUUGCAUCAUUUCAACUGCCCGGAAUCGCAGUGACUGUAGACUAAAU---UUUGGACCGAAGC------CUACAGCCCUGCAAGACAGCAGUUA ---((((((---........................))))))....((((.(.((((((.((....---..........)).------)))))).)))))............. ( -21.80, z-score = 0.58, R) >droYak2.chr2L 3063753 101 - 22324452 ---CCGGGCUCUCUUUCAUCUUGCAUCAUUUCAACUGCCCGGAAUCGCAGCGACUGUAGACUAAAU---UUUGGACCAAAGC------CUACAGCCCUGCAAGACAGCAGUUA ---((((((...........................))))))....((((.(.((((((.((....---..........)).------)))))).)))))............. ( -25.07, z-score = -0.76, R) >droEre2.scaffold_4929 3115497 98 - 26641161 ---CUGGGC---CUCUCAUCUUGCAUCCUUUCAACUGCCCGGAAUCGCAGCGACUGUAGACUAAAU---UUUGGACCGAAGC------CUACAGCCCUGCAAGACAGCAGUUA ---((((((---........................))))))....((((.(.((((((.((....---..........)).------)))))).)))))............. ( -21.80, z-score = 0.70, R) >droAna3.scaffold_12916 6592632 98 + 16180835 UUUUUGGGG---AGUCCGUAUUAUCAC-UUUCAACUGCCCGGAAUCGCAGUGGCUGUAGAC--AAA---UUUGGACCACGUC------CACAGACUCUGCAAGACAGCAGUUA .(((((.((---((((.((....((((-..(((.((((........)))))))..)).)).--...---...((((...)))------))).)))))).)))))......... ( -26.10, z-score = -0.67, R) >dp4.chr4_group3 4526508 95 + 11692001 ------------AAGGUGGAUCCCAUCCACUCAACUGCCCGGAAUCGCAGCGUCUGUAGACUGAAAU--UUUGGACCUAU-CGUC---CACAGGCGCUGCAAGACGGCAGUUA ------------..(((((((...))))))).(((((((.(.....(((((((((((.(((.((...--..........)-))))---.)))))))))))....)))))))). ( -40.02, z-score = -4.24, R) >droPer1.super_1 1669963 95 + 10282868 ------------AAGGUGGAUCCCAUCCACUCAACUGCCCGGAAUCGCAGCGUCUGUAGACUGAAAU--UUUGGACCUAU-CGUC---CACAGGCGCUGCAAGACGGCAGUUA ------------..(((((((...))))))).(((((((.(.....(((((((((((.(((.((...--..........)-))))---.)))))))))))....)))))))). ( -40.02, z-score = -4.24, R) >droWil1.scaffold_181132 407342 88 - 1035393 ------------------------UGUCAUUCAAUUGCCCAGAAUCGCAGGAUCUAUAAAGAA-UUUAAUUUGGACCUGGAUUCUUUACAAAGACCCUGCAAGACGGCAAUUA ------------------------........(((((((.......(((((.(((.(((((((-((((.........)))))))))))...))).))))).....))))))). ( -25.20, z-score = -3.46, R) >droVir3.scaffold_12963 5738683 87 + 20206255 --------------------UUUAGUAAAAUCAACUGCCCAGAAUCGCAACUCGUGUGGAUU-AAUU--UUUGGACCUUUGUAUC---CACAGGCGUUGCAAGACGGCAGUUA --------------------............(((((((.......(((((.(.((((((((-((..--.........))).)))---)))).).))))).....))))))). ( -27.50, z-score = -3.08, R) >droGri2.scaffold_15252 2029281 86 - 17193109 ------------------------CUGAGUUUAACUGCUCAGAAUCGCCGAGCAUGUGGGCAACUUUUGUUUGGACCUGGGUGCC---CACAGGCCCGGCAAGACGGCAGUUA ------------------------((((((......))))))....((((.((.((((((((.(((..((....))..)))))))---)))).)).))))..(((....))). ( -36.80, z-score = -2.81, R) >consensus _____G_G____CUCUCAUCUUGCAUCAUUUCAACUGCCCGGAAUCGCAGCGACUGUAGACUAAAU___UUUGGACCUAAGC______CAAAGGCCCUGCAAGACAGCAGUUA ................................((((((...(..(.((((..............................................)))).)..).)))))). ( -8.68 = -8.19 + -0.49)
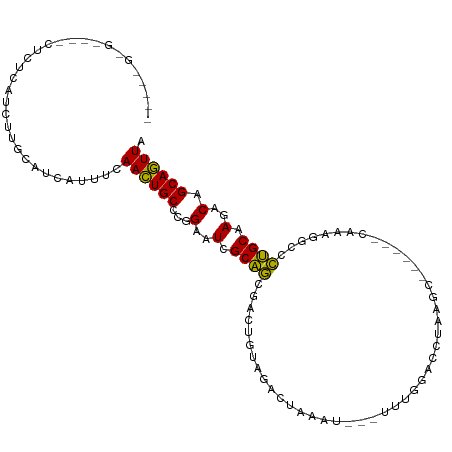
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:12:57 2011