
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,628,462 – 12,628,554 |
| Length | 92 |
| Max. P | 0.830270 |

| Location | 12,628,462 – 12,628,554 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 68.68 |
| Shannon entropy | 0.50244 |
| G+C content | 0.48701 |
| Mean single sequence MFE | -19.80 |
| Consensus MFE | -15.60 |
| Energy contribution | -15.92 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.21 |
| Mean z-score | -0.75 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.830270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12628462 92 - 21146708 --------UAUGUGCGUGUGUGUGUACAAGUGUAUGUACAUGCUAAAUCUCACUAUCCCACC-----------CCUUUGUUUCCCCCUGGCCAUAGCCUUAACAUUACGCA --------....((((((.(((((((((......)))))))))...................-----------....((((.......(((....)))..)))).)))))) ( -19.50, z-score = -1.37, R) >droSim1.chr2R 11363297 97 - 19596830 ------------UGUGUGCGUGUGUACAUGUGUAUGUACAUGCUAAAUCUCACCACCCCCCC--AUCGCCCACCCCUUUGUUCCCGCUGGCCAUAGCCUUAACAUUACGCA ------------.(((((.((((((((((....))))))))))...................--..............((((...((((....))))...)))).))))). ( -19.70, z-score = -0.87, R) >droSec1.super_1 10134019 98 - 14215200 ------------UGUGUGCGUGUGUACAUGUGUAUGUACAUGCUAAAUCUCACCACCCCCCCCUAUCGUCCACCCCCUU-UUCCCACUGGCCAUAGCCUUAACAUUACGCA ------------.(((((.((((((((((....))))))))))....................................-........(((....))).......))))). ( -18.40, z-score = -1.48, R) >droYak2.chr2R 4788710 91 + 21139217 UACCUAUGUACAUAUGUGCGUGUGUACAUGUGUAUGUACAUGCCAAAU----CUGCCCCA----------------UUCCUGGCCAGCCUCCAUAGCCUUAACAUUACGCA ....(((((((((((((((....)))))))))))))))..(((.....----..(((...----------------.....)))..((.......))...........))) ( -20.30, z-score = -0.30, R) >droEre2.scaffold_4845 9365571 79 + 22589142 --------CAUGUGCGUGUGCACGUACAUGUGUAUGUACAUGCCAAAU----ACACCCCA--------------------UUCCUGCUGGCCAUGGCUUUAACAUUACGCA --------.((((((((....))))))))((((((((....((((...----.((.(...--------------------.....).))....))))....))).))))). ( -21.10, z-score = 0.30, R) >consensus _________A__UGUGUGCGUGUGUACAUGUGUAUGUACAUGCUAAAUCUCACCACCCCACC___________CC_UUU_UUCCCACUGGCCAUAGCCUUAACAUUACGCA ............((((((.((((((((((....)))))))))).............................................(((....))).......)))))) (-15.60 = -15.92 + 0.32)

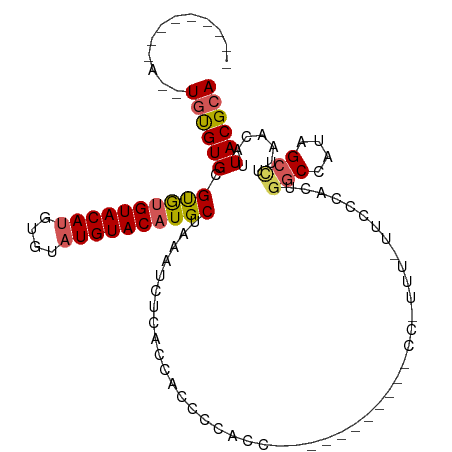

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:28:02 2011