
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,621,193 – 12,621,288 |
| Length | 95 |
| Max. P | 0.578198 |

| Location | 12,621,193 – 12,621,288 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.08 |
| Shannon entropy | 0.42945 |
| G+C content | 0.43471 |
| Mean single sequence MFE | -26.15 |
| Consensus MFE | -12.43 |
| Energy contribution | -12.67 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.578198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12621193 95 + 21146708 ----GCCAGUUACUUAGUUAGCCGGG-CUAAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCUCGCUU--UCCGUCUCAUUACAGACUGC-------------- ----(((((((.....((((((....-....)))))).....(((((((((....))))))))).)))))))........--...((((......))))...-------------- ( -27.40, z-score = -2.01, R) >droSim1.chr2R 11356138 95 + 19596830 ----GCCAGUUAGUUAGUUAGCCGGG-CUAAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCUCGCUU--UCCGUCUCAUUACAGACUGC-------------- ----(((((((.((((((((((...)-)).))))))).....(((((((((....))))))))).)))))))........--...((((......))))...-------------- ( -29.00, z-score = -2.25, R) >droSec1.super_1 10127042 95 + 14215200 ----GCCAGUUAGUUAGUUAGCCGGG-CUAAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCUCGCUU--UCCGUCUCAUUACAGACUGC-------------- ----(((((((.((((((((((...)-)).))))))).....(((((((((....))))))))).)))))))........--...((((......))))...-------------- ( -29.00, z-score = -2.25, R) >droYak2.chr2R 4780965 95 - 21139217 ----GCCAGUUUGUUAGUUAGCCGGG-CUAAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCUCGCUU--UCCGUCUCAUUAAAGACUGC-------------- ----((((((((((((((((((...)-)).))))))))))).(((((((((....)))))))))....))))........--...((((......))))...-------------- ( -29.70, z-score = -2.56, R) >droEre2.scaffold_4845 9357607 109 - 22589142 ----GCCAGUUAGUUAGUUAGCCGGG-CUAAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCUCGCUU--UCCGUCUCAUUAAAGACUGCAGGCCCCCAACUGC ----(((((((.((((((((((...)-)).))))))).....(((((((((....))))))))).)))))))........--...((((......)))).((((........)))) ( -33.50, z-score = -2.00, R) >droAna3.scaffold_13266 5908684 106 - 19884421 GCCAGUUGGUUAGUUAGUUAGCCGGG-CUAAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCUCGCUU--UCCGUCUCAUUACAGACUGUAGGCUGC------- (((((((((((((((((((.....))-)).))))))).....(((((((((....)))))))))))))))))((..((((--.(.((((......)))).).)))).))------- ( -36.60, z-score = -2.68, R) >dp4.chr3 686453 102 - 19779522 --------GCCUCCUGGUUAGCCGGAGCUAAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCUCGCUU--UCCGUCUCAUUAAAGAUUUAACUCUGGGCC---- --------((((...((..(((..((((...((..(......(((((((((....)))))))))...)..))))))))).--.))((((......))))........)))).---- ( -26.40, z-score = -0.80, R) >droPer1.super_37 664401 102 - 760358 --------GCCUCCUGGUUAGCCGGAGCUAAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCUCGCUU--UCCGUCUCAUUAAAGAUUUAACUCUGGGCC---- --------((((...((..(((..((((...((..(......(((((((((....)))))))))...)..))))))))).--.))((((......))))........)))).---- ( -26.40, z-score = -0.80, R) >droWil1.scaffold_180569 790836 90 - 1405142 --------GCCAGUUAGUCAAGCUA--CUAAGCUGACAAAUACUAUAAUUAAUUUUAAUUGCACCGAUUGGCGCUCUCUU--UCCGUCUCAUUAAAGUAUUU-------------- --------(((((((.((((.(((.--...)))))))........((((((....))))))....)))))))........--....................-------------- ( -17.20, z-score = -1.92, R) >droMoj3.scaffold_6496 22527266 88 + 26866924 ------------GCCAGUUAGGCCA--GAAAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCCCGCUUUUUCCGUCUCAUUAAAGUUUGC-------------- ------------(((((((.(((..--....)))........(((((((((....))))))))).)))))))((..(((((............)))))..))-------------- ( -20.90, z-score = -1.05, R) >droVir3.scaffold_12875 17910223 85 - 20611582 ------------GCCAGUUAG-CCA--CAAAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCCCGCUU--UCCGUCUCAUUAAGUUCUGC-------------- ------------(((((((((-(..--....)))))).....(((((((((....)))))))))....))))((..((((--...........))))...))-------------- ( -21.80, z-score = -2.16, R) >droGri2.scaffold_15112 3472170 73 + 5172618 ---------------------GCCA--GUUAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCCCGCUU--UCCGUCUCAUUAAAGG------------------ ---------------------((((--(((.((((.(((.((..((.....))..)).))))))))))))))........--................------------------ ( -15.90, z-score = -0.96, R) >consensus ________GUUAGUUAGUUAGCCGGG_CUAAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCUCGCUU__UCCGUCUCAUUAAAGACUGC______________ ..................................(((.....(((((((((....))))))))).((..(((....)))...)).)))............................ (-12.43 = -12.67 + 0.25)

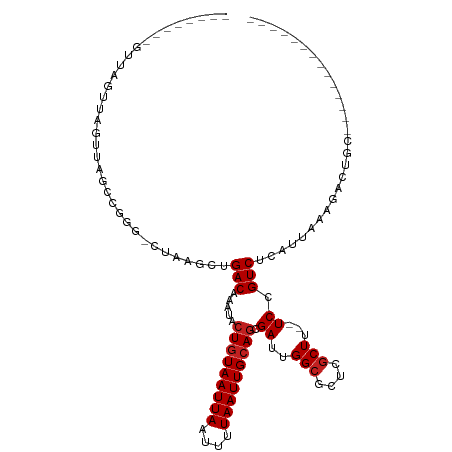
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:28:01 2011