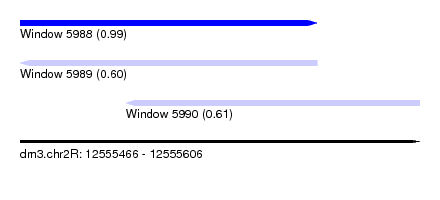
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,555,466 – 12,555,606 |
| Length | 140 |
| Max. P | 0.993283 |

| Location | 12,555,466 – 12,555,570 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 67.35 |
| Shannon entropy | 0.49793 |
| G+C content | 0.52702 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -17.16 |
| Energy contribution | -16.85 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.993283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12555466 104 + 21146708 GAAAAAAGAGUAACAGCCCGCUCUAUAGAAGAGAGUAUUCGCAAACCUGU-GUACUCCCGCAGCAAAUCG----------UGUGCCGCUCACGUGAGCACACUCGCACACUUUCU ......(((((........))))).....(((((((....((....((((-(......)))))......(----------((((((((....))).))))))..))..))))))) ( -29.00, z-score = -2.33, R) >droEre2.scaffold_4845 9284396 90 - 22589142 ----GAAAGGUGCCAGUCCGCUCUAUAG---AGGGUAUUCCCAUGCCU------GUACUCCCGCACAUCG----------UGUGCCGCUCACGUGAGCACUCUCGCACCUUCU-- ----..(((((((.((...((((....(---(((((((....))))).------........((((....----------.))))..)))....))))...)).)))))))..-- ( -29.90, z-score = -2.31, R) >droYak2.chr2R 4713452 107 - 21139217 ---GAAAGAGUGGCAUUCCGCUUUAUAG---AGAGUAUUCGUAUACCUAUAGCCGUACUCCCGCAAAUCGCCGCAAAUCGUGUGCCGCUCACGUGAGCACACUCGCACUUUCU-- ---(((((.(((((.....(((......---((.((((....))))))..))).((......)).....))))).....(((((((((....))).)))))).....))))).-- ( -30.60, z-score = -2.22, R) >droSec1.super_1 10053375 78 + 14215200 GAAAAGAGAGUGGCAGCCCGCUCUAUAG---AGAGUAUUCGCAUA----------------------UCG----------UGUGCCGCUCACGUGAGCACACUCGCACUUUCU-- ....((((((((.(.((..(((((....---)))))....))...----------------------..(----------((((((((....))).))))))..)))))))))-- ( -28.00, z-score = -2.50, R) >consensus ___AAAAGAGUGGCAGCCCGCUCUAUAG___AGAGUAUUCGCAUACCU______GUACUCCCGCAAAUCG__________UGUGCCGCUCACGUGAGCACACUCGCACUUUCU__ ....((((((((.......(((((.......))))).............................................(((((((....))).)))).....)))))))).. (-17.16 = -16.85 + -0.31)

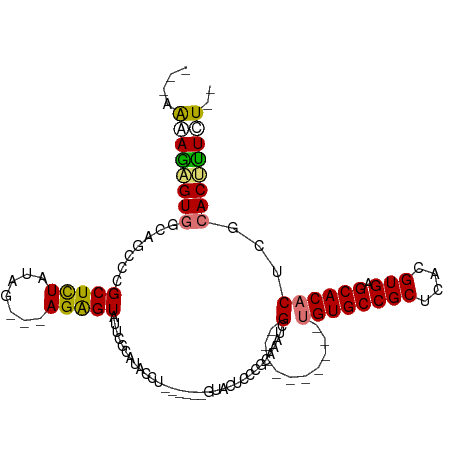
| Location | 12,555,466 – 12,555,570 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 67.35 |
| Shannon entropy | 0.49793 |
| G+C content | 0.52702 |
| Mean single sequence MFE | -32.08 |
| Consensus MFE | -15.86 |
| Energy contribution | -17.42 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.598482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12555466 104 - 21146708 AGAAAGUGUGCGAGUGUGCUCACGUGAGCGGCACA----------CGAUUUGCUGCGGGAGUAC-ACAGGUUUGCGAAUACUCUCUUCUAUAGAGCGGGCUGUUACUCUUUUUUC ((((((.(((..(((.(((((......((((((..----------.....))))))(((((((.-.(........)..))))))).......))))).)))..))).)))))).. ( -33.80, z-score = -1.01, R) >droEre2.scaffold_4845 9284396 90 + 22589142 --AGAAGGUGCGAGAGUGCUCACGUGAGCGGCACA----------CGAUGUGCGGGAGUAC------AGGCAUGGGAAUACCCU---CUAUAGAGCGGACUGGCACCUUUC---- --.((((((((.((.((((((.(....)(.((((.----------....)))).)))))))------.............((((---(....))).)).)).)))))))).---- ( -36.80, z-score = -3.00, R) >droYak2.chr2R 4713452 107 + 21139217 --AGAAAGUGCGAGUGUGCUCACGUGAGCGGCACACGAUUUGCGGCGAUUUGCGGGAGUACGGCUAUAGGUAUACGAAUACUCU---CUAUAAAGCGGAAUGCCACUCUUUC--- --.(((((.((((((((((.(.(....).)))))))...))))((((.((((((((((((((............))..))))))---)......))))).))))...)))))--- ( -35.20, z-score = -1.53, R) >droSec1.super_1 10053375 78 - 14215200 --AGAAAGUGCGAGUGUGCUCACGUGAGCGGCACA----------CGA----------------------UAUGCGAAUACUCU---CUAUAGAGCGGGCUGCCACUCUCUUUUC --(((.((((((.((((((.(.(....).))))))----------)..----------------------..)))....))).)---))..((((.((....)).))))...... ( -22.50, z-score = -0.14, R) >consensus __AGAAAGUGCGAGUGUGCUCACGUGAGCGGCACA__________CGAUUUGCGGGAGUAC______AGGUAUGCGAAUACUCU___CUAUAGAGCGGACUGCCACUCUUUU___ ..((((((((..(((.(((((..(((.....)))..................................(((((....)))))..........))))).)))..)))))))).... (-15.86 = -17.42 + 1.56)

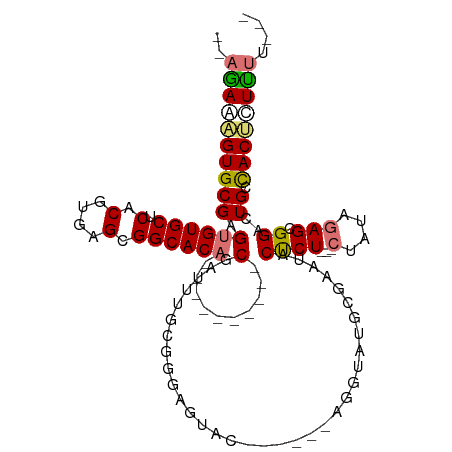
| Location | 12,555,503 – 12,555,606 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 65.98 |
| Shannon entropy | 0.57016 |
| G+C content | 0.49703 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -11.79 |
| Energy contribution | -12.33 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.611178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
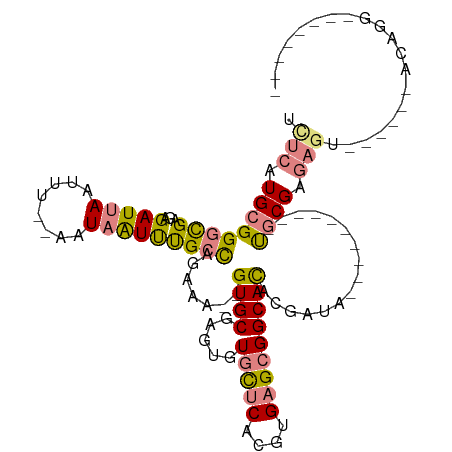
>dm3.chr2R 12555503 103 - 21146708 UCUCAUGCGGGCGAGAGAUUAAUUU--AAUAAUUUGCCAGAAAGUGUGCGAGUGUGCUCACGUGAGCGGCACACGAUUUGC-------UGCGGGAGUAC----ACAGGUUUGCGAA ((.(((((.((((((..((((...)--)))..)))))).....))))).))((((((((......((((((.......)))-------)))..))))))----))........... ( -33.00, z-score = -1.08, R) >droPer1.super_37 590915 90 + 760358 UUUCAUGCGGUUG-GAGAUAACUUUCGACU-GUCCGCUCUCA---GUGCUAGAAUUUGCACGUGAGCGGCAUGUUGAAGC-------GCACGAGAGA------ACAGC-------- ((((.((((((((-....))))((((((((-(.((((((...---((((........))))..)))))))).))))))))-------))).))))..------.....-------- ( -35.00, z-score = -2.77, R) >dp4.chr3 611076 90 + 19779522 UUUCAUGCGGUUG-GAGAUAACUUUCGACU-GUCCGCUCUCA---GUGCUAGAAUUUGCACGUGAGCGGCAUGUUGAAGA-------GCACGAGAGA------ACAGC-------- ((((.((((((((-....)))))(((((((-(.((((((...---((((........))))..)))))))).))))))..-------))).))))..------.....-------- ( -33.60, z-score = -2.68, R) >droEre2.scaffold_4845 9284426 96 + 22589142 UCUCAUGCGGGCGAGAGAUUAAGUU--AUUAAUUUGCCAGAAG--GUGCGAGAGUGCUCACGUGAGCGGCACACGAUG----------UGCGGGAGU------ACAGGCAUGGGAA ((((((((.(((((...(((((...--.)))))))))).....--........((((((.(....)(.((((.....)----------))).)))))------))..)))))))). ( -35.20, z-score = -3.15, R) >droYak2.chr2R 4713483 112 + 21139217 UCUCAUGCGGGCGAGAGAUUAAUUU--AUUAAUUUGCCAGAAA--GUGCGAGUGUGCUCACGUGAGCGGCACACGAUUUGCGGCGAUUUGCGGGAGUACGGCUAUAGGUAUACGAA ((..((((.(((((...(((((...--.))))))))))((...--((((..((((((.(.(....).)))))))..(((((((....))))))).))))..))....))))..)). ( -29.90, z-score = 0.47, R) >droSec1.super_1 10053409 80 - 14215200 UCUCAUGCGGGCGAGAGAUUAAUUU--AAUAAUUUGCCAGAAA--GUGCGAGUGUGCUCACGUGAGCGGCACACGAUA----------UGCGAA---------------------- ((.(((((.((((((..((((...)--)))..)))))).....--......((((((.(.(....).)))))))).))----------)).)).---------------------- ( -23.50, z-score = -1.06, R) >droSim1.chr2R 11288928 82 - 19596830 UCUCAUGCGGGCGAGAGAUUAAUUUAGAAUAAUUUGCCAGAAA--GUGCGAGUGUGCUCACGUGAGCGGCACACGAUA----------UGCGAA---------------------- ((.(((((.((((((..(((.......)))..)))))).....--......((((((.(.(....).)))))))).))----------)).)).---------------------- ( -23.10, z-score = -0.81, R) >consensus UCUCAUGCGGGCGAGAGAUUAAUUU__AAUAAUUUGCCAGAAA__GUGCGAGUGUGCUCACGUGAGCGGCACACGAUA__________UGCGAGAGU______ACAGG________ .(((.((((((((...(((((........))))))))).......((((.....(((((....)))))))))................)))).))).................... (-11.79 = -12.33 + 0.54)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:27:52 2011