
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,548,278 – 12,548,387 |
| Length | 109 |
| Max. P | 0.704311 |

| Location | 12,548,278 – 12,548,387 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 69.84 |
| Shannon entropy | 0.56312 |
| G+C content | 0.42125 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -7.14 |
| Energy contribution | -9.22 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.704311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12548278 109 + 21146708 GCUGCGUAACGUCACGUAGUUACGCAGUUACGUAA-GCAACUUCAGUAACGACAUUUACUGGUAUUUCAAGAUGGAAGAUCGGCUUUCUUCUCCCAUUAAGUUGUAAAUA ((((((((((........)))))))))).......-(((((((((((((......)))))).........(((((.(((..........))).))))))))))))..... ( -32.80, z-score = -3.55, R) >droSim1.chr2R 11281703 99 + 19596830 GCUGCGUAACGUCAC--------GCAGUUACGUAA-GCAACUUCAGUAACGACUUUUACUGGGAUUUCAA--UGGAAGAUCGGCUUUCUUCUCCCAUUAAGUUGAAAAUA ((((((((((.....--------...))))))).)-))...........((((((....(((((......--..(((((.......))))))))))..))))))...... ( -27.20, z-score = -2.56, R) >droSec1.super_1 10046220 107 + 14215200 GCUGCGUAACGUCACACGGUUACGCAGUUACGUAA-GCAACUUCAGUAACGAAUUUUACUGGGAUUUCAA--UGGAAGAUCGGCUUUCUUCUCCCAUUAAGUUGUAAAUA ((((((((((........)))))))))).......-(((((((((((((......)))))).......((--(((.(((..........))).))))))))))))..... ( -31.80, z-score = -3.45, R) >droYak2.chr2R 4706199 106 - 21139217 GCUGCGUAACGUCACGCAGUUACGCAGUGACGUAC-GUAGGAACAGUAACAACGUA-AAUGGGAUUUCAA--UGGAAGUACGGCUUUCUUCUCCCAUUGAGUCGUAAAUA .(((((((.((((((((......)).)))))))))-))))...........(((..-(((((((......--.(((((.....)))))...)))))))....)))..... ( -35.00, z-score = -2.91, R) >droEre2.scaffold_4845 9277269 92 - 22589142 GCUGCGUAACGUCAU--------GCAGUUACGCA--GUGAC----GUAAGAACUUU--CUGGGAUUUCAA--UGGAAGUACGGCUUUCUUCUCCCCCUAAGUUGUAAAUA ((((((((((.....--------...))))))))--))...----...(.(((((.--..((((......--.(((((.....)))))...))))...))))).)..... ( -27.80, z-score = -2.51, R) >droAna3.scaffold_13266 5840496 85 - 19884421 -----------------------CCAGCAACGUAACGUAAGAGCCAGGGCCAAGGCAUAUGGAAUUUCAU--UGAAACUUUUGCUUUCGUCUGGCAUUAAAUGAUAAUAA -----------------------((((..(((.((.((((((((((..((....))...)))..((((..--.))))))))))).)))))))))................ ( -17.70, z-score = -0.32, R) >consensus GCUGCGUAACGUCAC________GCAGUUACGUAA_GCAACUUCAGUAACGACUUUUACUGGGAUUUCAA__UGGAAGAUCGGCUUUCUUCUCCCAUUAAGUUGUAAAUA ..((((((((................)))))))).........................(((((.........(((((.....)))))...))))).............. ( -7.14 = -9.22 + 2.09)

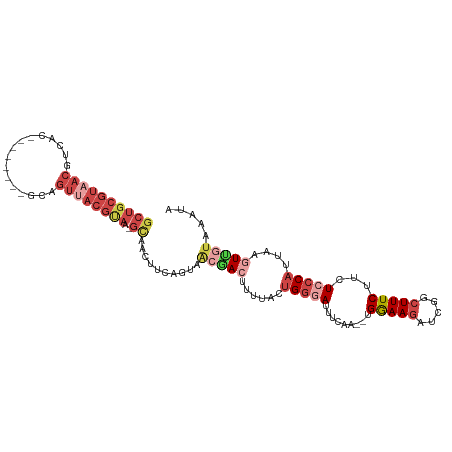
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:27:50 2011