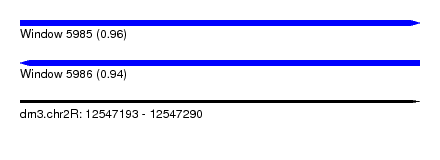
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,547,193 – 12,547,290 |
| Length | 97 |
| Max. P | 0.956140 |

| Location | 12,547,193 – 12,547,290 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 65.49 |
| Shannon entropy | 0.63900 |
| G+C content | 0.42842 |
| Mean single sequence MFE | -22.82 |
| Consensus MFE | -7.79 |
| Energy contribution | -7.60 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
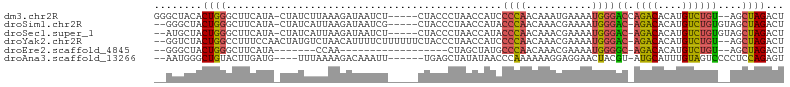
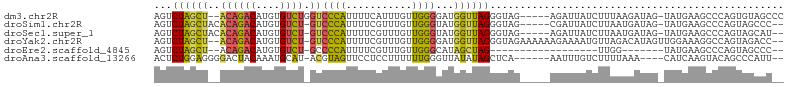
>dm3.chr2R 12547193 97 + 21146708 GGGCUACACUGGGCUUCAUA-CUAUCUUAAAGAUAAUCU-----CUACCCUAACCAUCCCCAACAAAUGAAAAUGGGACCAGACACAUGUCUGU--AGCUAGACU .((((((...(((.......-.((((.....))))....-----...)))........((((...........))))...((((....))))))--))))..... ( -20.16, z-score = -0.98, R) >droSim1.chr2R 11280612 96 + 19596830 --GGGCUACUGGGCUUCAUA-CUAUCAUUAAGAUAAUCG-----CUACCCUAACCAUACCCAACAAACGAAAAUGGGAC-AGACACAUGUCUGUGUAGCUAGACU --.((((((.(((.......-.((((.....))))....-----...)))........((((...........))))((-((((....))))))))))))..... ( -24.06, z-score = -2.27, R) >droSec1.super_1 10045111 96 + 14215200 --AUGCUACUGGGCUUCAUA-CUAUCAUUAAGAUAAUCU-----CUACCCUAACCAUACCCAACAAACGAAAAUGGGAC-AGACACAUGUCUGUGUAGCUAGACU --..(((((.(((.......-.((((.....))))....-----...)))........((((...........))))((-((((....)))))))))))...... ( -22.56, z-score = -2.57, R) >droYak2.chr2R 4705109 100 - 21139217 --GGUCUACUGGCCUUUCCAACUAUGUCUAACAUUUUCUUUUUUCUACCCUAACCAUCCCCAACAAACGAAAAUGGGAC-AGACACAUGUCUGU--AGCUAGACU --((((....))))...........(((((.(..........................((((...........))))((-((((....))))))--.).))))). ( -21.90, z-score = -2.56, R) >droEre2.scaffold_4845 9276227 75 - 22589142 --GGGCUACUGGGCUUCAUA-------CCAA------------------CUAGCUAUGCCCAACAAACGAAAAUGGGGC-AGACACAUGUCUGU--AGCUAGACU --(((((....)))))....-------....------------------((((((((((((..............))))-((((....))))))--))))))... ( -23.44, z-score = -2.33, R) >droAna3.scaffold_13266 5839443 92 - 19884421 --AAUGGGCUGUACUUGAUG----UUUAAAAGACAAAUU------UGAGCUAUAUAACCCAAAAAAGGAGGAACUACGU-AUGCAUUUGUAGUCCCCUCCAGAGU --..((((.((((((..(((----((.....))))...)------..))...)))).)))).....(((((.((((((.-.......))))))..)))))..... ( -24.80, z-score = -2.85, R) >consensus __GGGCUACUGGGCUUCAUA_CUAUCUUUAAGAUAAUCU_____CUACCCUAACCAUCCCCAACAAACGAAAAUGGGAC_AGACACAUGUCUGU__AGCUAGACU ........((((..............................................((((...........))))((.((((....))))))....))))... ( -7.79 = -7.60 + -0.19)
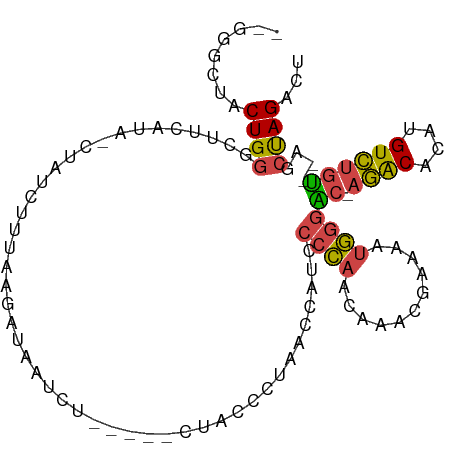
| Location | 12,547,193 – 12,547,290 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 65.49 |
| Shannon entropy | 0.63900 |
| G+C content | 0.42842 |
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -9.45 |
| Energy contribution | -8.82 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.57 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
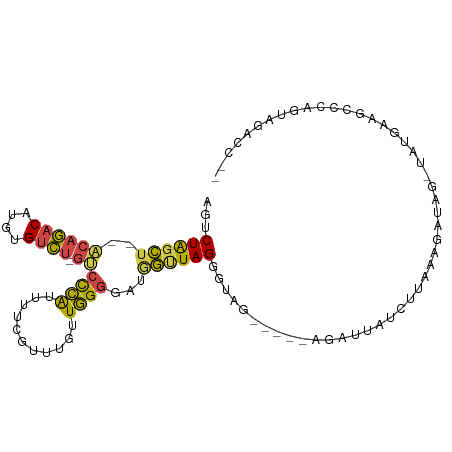
>dm3.chr2R 12547193 97 - 21146708 AGUCUAGCU--ACAGACAUGUGUCUGGUCCCAUUUUCAUUUGUUGGGGAUGGUUAGGGUAG-----AGAUUAUCUUUAAGAUAG-UAUGAAGCCCAGUGUAGCCC ......(((--((((((....))))....(((((..((.....))..)))))...((((..-----..((((((.....)))))-).....))))...))))).. ( -30.60, z-score = -2.30, R) >droSim1.chr2R 11280612 96 - 19596830 AGUCUAGCUACACAGACAUGUGUCU-GUCCCAUUUUCGUUUGUUGGGUAUGGUUAGGGUAG-----CGAUUAUCUUAAUGAUAG-UAUGAAGCCCAGUAGCCC-- ......(((((((((((....))))-)).((((.((((.....)))).))))...((((..-----((((((((.....)))))-).))..)))).)))))..-- ( -31.00, z-score = -2.93, R) >droSec1.super_1 10045111 96 - 14215200 AGUCUAGCUACACAGACAUGUGUCU-GUCCCAUUUUCGUUUGUUGGGUAUGGUUAGGGUAG-----AGAUUAUCUUAAUGAUAG-UAUGAAGCCCAGUAGCAU-- ......(((((((((((....))))-)).((((.((((.....)))).))))...((((..-----..((((((.....)))))-).....)))).)))))..-- ( -30.50, z-score = -2.92, R) >droYak2.chr2R 4705109 100 + 21139217 AGUCUAGCU--ACAGACAUGUGUCU-GUCCCAUUUUCGUUUGUUGGGGAUGGUUAGGGUAGAAAAAAGAAAAUGUUAGACAUAGUUGGAAAGGCCAGUAGACC-- .(((((((.--((((((....))))-)).(((((..((.....))..))))).....................)))))))....((((.....))))......-- ( -27.70, z-score = -2.07, R) >droEre2.scaffold_4845 9276227 75 + 22589142 AGUCUAGCU--ACAGACAUGUGUCU-GCCCCAUUUUCGUUUGUUGGGCAUAGCUAG------------------UUGG-------UAUGAAGCCCAGUAGCCC-- ...((((((--(.((((....))))-((((.((........)).)))).)))))))------------------((((-------........))))......-- ( -21.00, z-score = -1.18, R) >droAna3.scaffold_13266 5839443 92 + 19884421 ACUCUGGAGGGGACUACAAAUGCAU-ACGUAGUUCCUCCUUUUUUGGGUUAUAUAGCUCA------AAUUUGUCUUUUAAA----CAUCAAGUACAGCCCAUU-- .....((((((.(((((........-..)))))))))))...((((((((....))))))------)).(((((((.....----....))).))))......-- ( -20.30, z-score = -1.31, R) >consensus AGUCUAGCU__ACAGACAUGUGUCU_GUCCCAUUUUCGUUUGUUGGGGAUGGUUAGGGUAG_____AGAUUAUCUUAAAGAUAG_UAUGAAGCCCAGUAGACC__ ...((((((....((((....))))...((((...........))))...))))))................................................. ( -9.45 = -8.82 + -0.63)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:27:49 2011