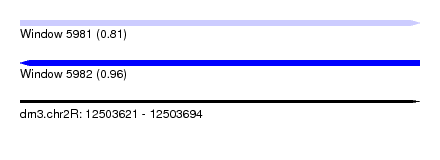
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,503,621 – 12,503,694 |
| Length | 73 |
| Max. P | 0.959546 |

| Location | 12,503,621 – 12,503,694 |
|---|---|
| Length | 73 |
| Sequences | 5 |
| Columns | 73 |
| Reading direction | forward |
| Mean pairwise identity | 84.35 |
| Shannon entropy | 0.27853 |
| G+C content | 0.52829 |
| Mean single sequence MFE | -23.05 |
| Consensus MFE | -19.47 |
| Energy contribution | -19.63 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.814061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
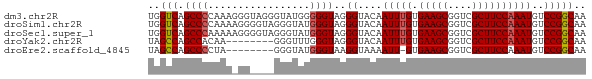
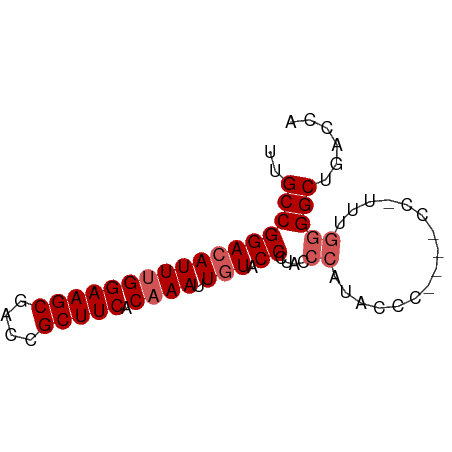
>dm3.chr2R 12503621 73 + 21146708 UGGUCAGCCCCAAAGGGUAGGGUAUGGGGGUAGGGUACAAUUUGUGAAGCGGUCGCUUCCAAAUGUCCGGCAA ..(((.(((((...............)))))..((.(((.((((.(((((....))))))))))))))))).. ( -24.26, z-score = -1.11, R) >droSim1.chr2R 11240796 73 + 19596830 UGGUCAGCCCCAAAAGGGGUAGGGUAUGGGUAGGGUACAAUUUGUGAAGCGGUCGCUUCCAAAUGUCCGGCAA ..(((.(((((....))))).............((.(((.((((.(((((....))))))))))))))))).. ( -25.70, z-score = -1.89, R) >droSec1.super_1 10001651 73 + 14215200 UGGUCAGCCCAAAAAGGGGUAGGGUAUGGGUAGGGUACAAUUUGUGAAGCGGUCGCUUCCAAAUGUCCGGCAA ..(((.((((......)))).............((.(((.((((.(((((....))))))))))))))))).. ( -22.50, z-score = -1.31, R) >droYak2.chr2R 4660941 65 - 21139217 UAGCCAGCCACAA--------GGGUUUGGGUAGGGUACAAUUUGUGAAGCGGUCGCUUCCAAAUGUCCGGCAA ..(((.(((.(((--------....))))))..((.(((.((((.(((((....))))))))))))))))).. ( -22.80, z-score = -1.99, R) >droEre2.scaffold_4845 9232867 64 - 22589142 UAGCCAGCCCCUA--------GGGUAUGGGUAAGGUAAAAUU-GUGAAGCGGUCGCUUCCAAAUGUCCGGCAA ..(((.((((...--------......))))..((...(.((-(.(((((....)))))))).)..))))).. ( -20.00, z-score = -1.02, R) >consensus UGGUCAGCCCCAAA_GG____GGGUAUGGGUAGGGUACAAUUUGUGAAGCGGUCGCUUCCAAAUGUCCGGCAA ..(((.((((.................))))..((....(((((.(((((....))))))))))..))))).. (-19.47 = -19.63 + 0.16)
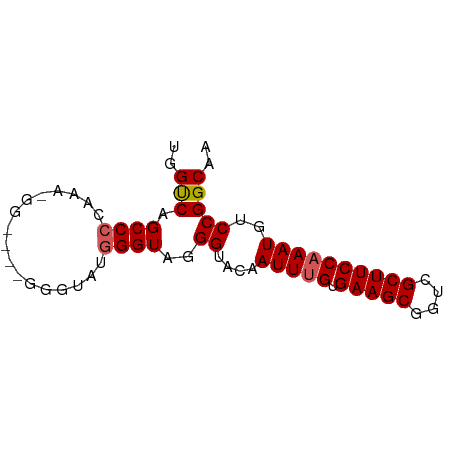
| Location | 12,503,621 – 12,503,694 |
|---|---|
| Length | 73 |
| Sequences | 5 |
| Columns | 73 |
| Reading direction | reverse |
| Mean pairwise identity | 84.35 |
| Shannon entropy | 0.27853 |
| G+C content | 0.52829 |
| Mean single sequence MFE | -19.79 |
| Consensus MFE | -13.35 |
| Energy contribution | -14.15 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12503621 73 - 21146708 UUGCCGGACAUUUGGAAGCGACCGCUUCACAAAUUGUACCCUACCCCCAUACCCUACCCUUUGGGGCUGACCA .....((((((((((((((....))))).)))).))).))....(((((............)))))....... ( -20.30, z-score = -2.61, R) >droSim1.chr2R 11240796 73 - 19596830 UUGCCGGACAUUUGGAAGCGACCGCUUCACAAAUUGUACCCUACCCAUACCCUACCCCUUUUGGGGCUGACCA .....((((((((((((((....))))).)))).))).))...........(..((((....))))..).... ( -20.60, z-score = -2.81, R) >droSec1.super_1 10001651 73 - 14215200 UUGCCGGACAUUUGGAAGCGACCGCUUCACAAAUUGUACCCUACCCAUACCCUACCCCUUUUUGGGCUGACCA .....((((((((((((((....))))).)))).))).))(..((((...............))))..).... ( -17.16, z-score = -2.27, R) >droYak2.chr2R 4660941 65 + 21139217 UUGCCGGACAUUUGGAAGCGACCGCUUCACAAAUUGUACCCUACCCAAACCC--------UUGUGGCUGGCUA ..(((((((((((((((((....))))).)))).)))......(((((....--------))).))))))).. ( -20.50, z-score = -2.80, R) >droEre2.scaffold_4845 9232867 64 + 22589142 UUGCCGGACAUUUGGAAGCGACCGCUUCAC-AAUUUUACCUUACCCAUACCC--------UAGGGGCUGGCUA ..(((((.(..((((((((....))))).)-))..........(((......--------..))))))))).. ( -20.40, z-score = -2.53, R) >consensus UUGCCGGACAUUUGGAAGCGACCGCUUCACAAAUUGUACCCUACCCAUACCC____CC_UUUGGGGCUGACCA ..(((((((((((((((((....))))).)))).))).))....((................)))))...... (-13.35 = -14.15 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:27:45 2011