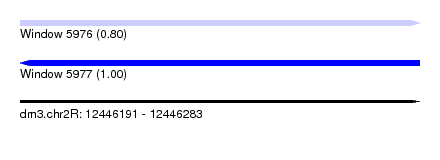
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,446,191 – 12,446,283 |
| Length | 92 |
| Max. P | 0.997617 |

| Location | 12,446,191 – 12,446,283 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 75.15 |
| Shannon entropy | 0.46992 |
| G+C content | 0.35679 |
| Mean single sequence MFE | -17.80 |
| Consensus MFE | -11.06 |
| Energy contribution | -10.28 |
| Covariance contribution | -0.78 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.801812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
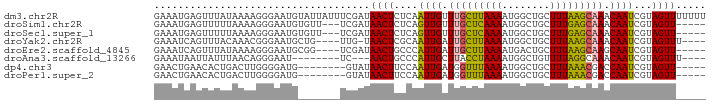
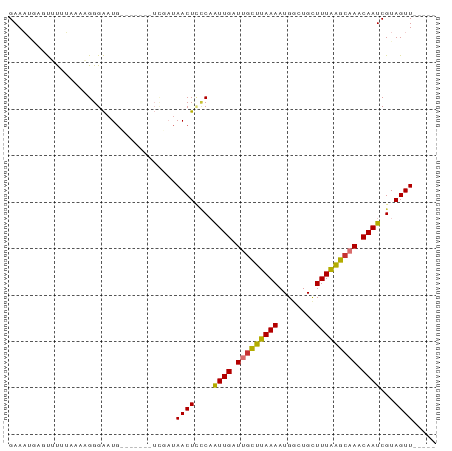
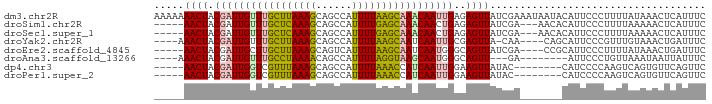
>dm3.chr2R 12446191 92 + 21146708 GAAAUGAGUUUAUAAAAGGGAAUGUAUUAUUUCGAUAACUCUCAAUUGUUUGCUUAAAAUGGCUGCUUUAAGCAAACAAUCGUAGUUUUUUU (((((((...(((........)))..)))))))...((((..(.((((((((((((((........)))))))))))))).).))))..... ( -20.80, z-score = -2.53, R) >droSim1.chr2R 11180149 84 + 19596830 GAAAUGAGUUUUUAAAAGGGAAUGUGUU---UCGAUAACUCUCAGUUGUUUGCUCAAAAUGGCUGCUUUGAGCAAACAAUCGUAGUU----- ((((((.((((((....)))))).))))---))...((((..(.((((((((((((((........)))))))))))))).).))))----- ( -23.70, z-score = -2.98, R) >droSec1.super_1 9944282 84 + 14215200 GAAAUGAGUUUUUAAAAGGGAAUGUGUU---UCGAUAACUCUCAGUUGUUUGCUCAAAAUGGCUGCUUUGAGCAAACAAUCGUAGUU----- ((((((.((((((....)))))).))))---))...((((..(.((((((((((((((........)))))))))))))).).))))----- ( -23.70, z-score = -2.98, R) >droYak2.chr2R 4599781 83 - 21139217 GAAAUCAGUUUACAAACGGGAAUGCUG----UUG-UAACUCGCAAUUGAUUGCUUAAAAUGGCUGCUUUAAGCAAACAAUCGUAGUUU---- ..........(((((.(((.....)))----)))-))(((.((.((((.(((((((((........))))))))).)))).)))))..---- ( -17.90, z-score = -1.01, R) >droEre2.scaffold_4845 9173760 83 - 22589142 GAAAUCAGUUUAUAAAAGGGAAUGCGG----UCGAUAACUGCCCAUUGAUUGCUUAAAAUGACUGCUUUAAGCAAGCAAUCGUAGUU----- ((.....((((........))))....----))...((((((..((((.(((((((((........))))))))).)))).))))))----- ( -16.10, z-score = -0.32, R) >droAna3.scaffold_13266 15814729 77 + 19884421 GAAAUAAUUAUUUAACAGGGAAU--------UC---AACUGCCCAUUGCUUACCUAAAAUGGCUGUUUUAGGCAAACAAUCGUAGUUU---- .......................--------..---((((((..((((.((.((((((((....)))))))).)).)))).)))))).---- ( -14.00, z-score = -0.79, R) >dp4.chr3 16080928 79 - 19779522 GAACUGAACACUGACUUGGGGAUG--------GUAUAACUUCCAAUUGAUGGUUUAAAAUGGCUGCUUUAAACGACCAAUCGUAGUU----- .........((((..(((((((.(--------......).)))........(((((((........)))))))..))))...)))).----- ( -13.10, z-score = 0.06, R) >droPer1.super_2 5151416 79 - 9036312 GAACUGAACACUGACUUGGGGAUG--------GUAUAACUUCCAAUUGAUGGUUUAAAAUGGCUGCUUUAAACGACCAAUCGUAGUU----- .........((((..(((((((.(--------......).)))........(((((((........)))))))..))))...)))).----- ( -13.10, z-score = 0.06, R) >consensus GAAAUGAGUUUUUAAAAGGGAAUG_______UCGAUAACUCCCAAUUGAUUGCUUAAAAUGGCUGCUUUAAGCAAACAAUCGUAGUU_____ ....................................((((....((((.(((((((((........))))))))).))))...))))..... (-11.06 = -10.28 + -0.78)
| Location | 12,446,191 – 12,446,283 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 75.15 |
| Shannon entropy | 0.46992 |
| G+C content | 0.35679 |
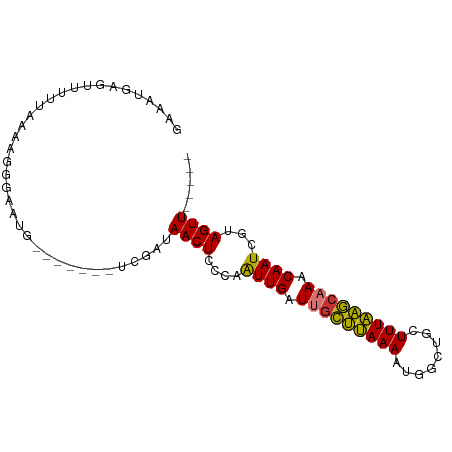
| Mean single sequence MFE | -18.61 |
| Consensus MFE | -18.82 |
| Energy contribution | -18.34 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.13 |
| Structure conservation index | 1.01 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.997617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
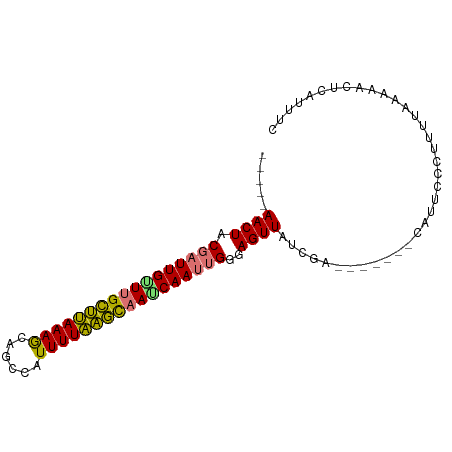
>dm3.chr2R 12446191 92 - 21146708 AAAAAAACUACGAUUGUUUGCUUAAAGCAGCCAUUUUAAGCAAACAAUUGAGAGUUAUCGAAAUAAUACAUUCCCUUUUAUAAACUCAUUUC ..........(((((((((((((((((......))))))))))))))))).(((((...((((............))))...)))))..... ( -22.10, z-score = -5.14, R) >droSim1.chr2R 11180149 84 - 19596830 -----AACUACGAUUGUUUGCUCAAAGCAGCCAUUUUGAGCAAACAACUGAGAGUUAUCGA---AACACAUUCCCUUUUAAAAACUCAUUUC -----.....((.((((((((((((((......)))))))))))))).)).(((((.(..(---((.........)))..).)))))..... ( -21.40, z-score = -4.49, R) >droSec1.super_1 9944282 84 - 14215200 -----AACUACGAUUGUUUGCUCAAAGCAGCCAUUUUGAGCAAACAACUGAGAGUUAUCGA---AACACAUUCCCUUUUAAAAACUCAUUUC -----.....((.((((((((((((((......)))))))))))))).)).(((((.(..(---((.........)))..).)))))..... ( -21.40, z-score = -4.49, R) >droYak2.chr2R 4599781 83 + 21139217 ----AAACUACGAUUGUUUGCUUAAAGCAGCCAUUUUAAGCAAUCAAUUGCGAGUUA-CAA----CAGCAUUCCCGUUUGUAAACUGAUUUC ----......((((((.((((((((((......)))))))))).))))))((..(((-(((----..((......))))))))..))..... ( -18.00, z-score = -2.16, R) >droEre2.scaffold_4845 9173760 83 + 22589142 -----AACUACGAUUGCUUGCUUAAAGCAGUCAUUUUAAGCAAUCAAUGGGCAGUUAUCGA----CCGCAUUCCCUUUUAUAAACUGAUUUC -----((((.(.((((.((((((((((......)))))))))).)))).)..)))).....----........................... ( -15.80, z-score = -1.34, R) >droAna3.scaffold_13266 15814729 77 - 19884421 ----AAACUACGAUUGUUUGCCUAAAACAGCCAUUUUAGGUAAGCAAUGGGCAGUU---GA--------AUUCCCUGUUAAAUAAUUAUUUC ----.((((.(.(((((((((((((((......))))))))))))))).)..))))---..--------....................... ( -21.00, z-score = -3.34, R) >dp4.chr3 16080928 79 + 19779522 -----AACUACGAUUGGUCGUUUAAAGCAGCCAUUUUAAACCAUCAAUUGGAAGUUAUAC--------CAUCCCCAAGUCAGUGUUCAGUUC -----((((.((((((((.((((((((......)))))))).))))))))..))))....--------........................ ( -14.60, z-score = -2.05, R) >droPer1.super_2 5151416 79 + 9036312 -----AACUACGAUUGGUCGUUUAAAGCAGCCAUUUUAAACCAUCAAUUGGAAGUUAUAC--------CAUCCCCAAGUCAGUGUUCAGUUC -----((((.((((((((.((((((((......)))))))).))))))))..))))....--------........................ ( -14.60, z-score = -2.05, R) >consensus _____AACUACGAUUGUUUGCUUAAAGCAGCCAUUUUAAGCAAUCAAUUGGGAGUUAUCGA_______CAUUCCCUUUUAAAAACUCAUUUC .....((((.(((((((((((((((((......)))))))))))))))))..)))).................................... (-18.82 = -18.34 + -0.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:27:41 2011