
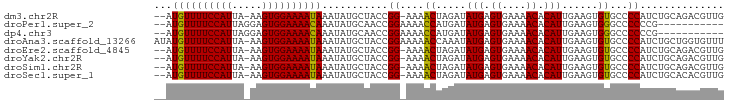
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,441,272 – 12,441,367 |
| Length | 95 |
| Max. P | 0.996694 |

| Location | 12,441,272 – 12,441,363 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 87.22 |
| Shannon entropy | 0.24588 |
| G+C content | 0.38411 |
| Mean single sequence MFE | -20.45 |
| Consensus MFE | -16.00 |
| Energy contribution | -15.44 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12441272 91 - 21146708 --AUGUUUUCCAUUA-AAGUGGAAAAUAAAUAUGCUACCGG-AAAACUAGAUAUGAGUGAAAACACAUUGAAGUGUGCCCCAUCUGCAGACGUUG --.((((((((((..-..))))))))))....(((....((-...(((....(((.((....)).)))...)))...))......)))....... ( -19.20, z-score = -1.98, R) >droPer1.super_2 5146628 82 + 9036312 --AUGUUUUCCAUUAGGAGUGGAAAACAAAUAUGCAACCGGAAAACCAUGAUAUGAGUGAAAACACAUUGAAGUGGGCCCCCCG----------- --.((((((((((.....))))))))))..........(((....((((...(((.((....)).)))....)))).....)))----------- ( -22.80, z-score = -2.55, R) >dp4.chr3 16076140 82 + 19779522 --AUGUUUUCCAUUAGGAGUGGAAAACAAAUAUGCAACCGGAAAACCAUGAUAUGAGUGAAAACACAUUGAAGUGGGCCCCCCG----------- --.((((((((((.....))))))))))..........(((....((((...(((.((....)).)))....)))).....)))----------- ( -22.80, z-score = -2.55, R) >droAna3.scaffold_13266 15810044 94 - 19884421 AUAUGUUUUCCAUUA-AAGUGGAAAAUAAAUAUGCUACCGGAAAAACCAAAUAUGAGUGAAAACACAUUGAAGUGUGCCCCAUCUGCUGGUGUUU ...((((((((((..-..))))))))))...((((((.((((...((((....)).)).....(((((....))))).....)))).)))))).. ( -19.90, z-score = -1.74, R) >droEre2.scaffold_4845 9168911 91 + 22589142 --AUGUUUUCCAUUA-AAGUGGAAAAUAAAUAUGCUACCGG-AAAACUAGAUAUGAGUGAAAACACAUUGAAGUGUGCCCCAUCUGCAGACGUUG --.((((((((((..-..))))))))))....(((....((-...(((....(((.((....)).)))...)))...))......)))....... ( -19.20, z-score = -1.98, R) >droYak2.chr2R 4594862 91 + 21139217 --AUGUUUUCCAUUA-AAGUGGAAAAUAAAUAUGCUACCGG-AAAACUAGAUAUGAGUGAAAACACAUUGAAGUGUGCCCCAUCUGCAGACGUUG --.((((((((((..-..))))))))))....(((....((-...(((....(((.((....)).)))...)))...))......)))....... ( -19.20, z-score = -1.98, R) >droSim1.chr2R 11174956 91 - 19596830 --AUGUUUUCCAUUA-AAGUGGAAAAUAAAUAUGCUACCGG-AAAACUAGAUAUGAGUGAAAACACAUUGAAGUGUGCCCCAUCUGCAGACGUUG --.((((((((((..-..))))))))))....(((....((-...(((....(((.((....)).)))...)))...))......)))....... ( -19.20, z-score = -1.98, R) >droSec1.super_1 9939451 91 - 14215200 --AUGUUUUCCAUUA-AAGUGGAAAAUAAAUAUGCUACCGG-AAAACUAGAUAUGAGUGAAAACACAUUGAAGUGUGCCCCAUCUGCACACGUUG --.((((((((((..-..)))))))))).............-..(((.....(((.((....)).)))....((((((.......))))))))). ( -21.30, z-score = -2.56, R) >consensus __AUGUUUUCCAUUA_AAGUGGAAAAUAAAUAUGCUACCGG_AAAACUAGAUAUGAGUGAAAACACAUUGAAGUGUGCCCCAUCUGCAGACGUUG ...((((((((((.....))))))))))...........((....((.....(((.((....)).)))......))...)).............. (-16.00 = -15.44 + -0.56)
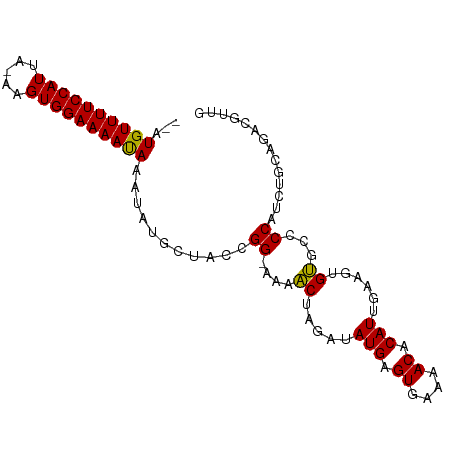
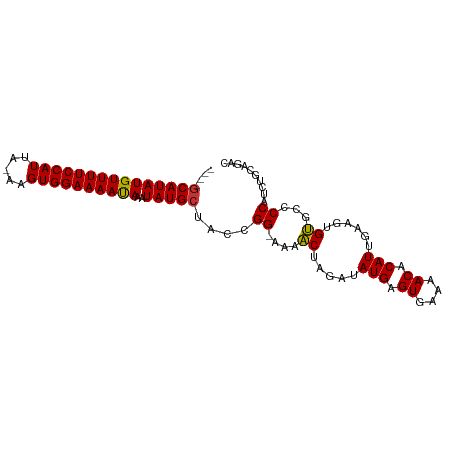
| Location | 12,441,276 – 12,441,367 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 88.53 |
| Shannon entropy | 0.21914 |
| G+C content | 0.38150 |
| Mean single sequence MFE | -23.16 |
| Consensus MFE | -19.86 |
| Energy contribution | -19.43 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.996694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12441276 91 - 21146708 ----GCAUAUGUUUUCCAUUA-AAGUGGAAAAUAAAUAUGCUACCGG-AAAACUAGAUAUGAGUGAAAACACAUUGAAGUGUGCCCCAUCUGCAGAC ----(((((((((((((((..-..)))))))))..))))))....((-...(((....(((.((....)).)))...)))...))............ ( -21.80, z-score = -2.99, R) >droPer1.super_2 5146631 83 + 9036312 ----GCAUAUGUUUUCCAUUAGGAGUGGAAAACAAAUAUGCAACCGGAAAACCAUGAUAUGAGUGAAAACACAUUGAAGUGGGCCCC---------- ----(((((((((((((((.....))))))))))..)))))....((....((((...(((.((....)).)))....)))).))..---------- ( -27.10, z-score = -4.25, R) >dp4.chr3 16076143 83 + 19779522 ----GCAUAUGUUUUCCAUUAGGAGUGGAAAACAAAUAUGCAACCGGAAAACCAUGAUAUGAGUGAAAACACAUUGAAGUGGGCCCC---------- ----(((((((((((((((.....))))))))))..)))))....((....((((...(((.((....)).)))....)))).))..---------- ( -27.10, z-score = -4.25, R) >droAna3.scaffold_13266 15810048 96 - 19884421 ACAUCCAUAUGUUUUCCAUUA-AAGUGGAAAAUAAAUAUGCUACCGGAAAAACCAAAUAUGAGUGAAAACACAUUGAAGUGUGCCCCAUCUGCUGGU .((..((((((((((((((..-..)))))))))............((.....))..)))))..))....(((((....)))))..(((.....))). ( -20.00, z-score = -1.50, R) >droEre2.scaffold_4845 9168915 91 + 22589142 ----GCAUAUGUUUUCCAUUA-AAGUGGAAAAUAAAUAUGCUACCGG-AAAACUAGAUAUGAGUGAAAACACAUUGAAGUGUGCCCCAUCUGCAGAC ----(((((((((((((((..-..)))))))))..))))))....((-...(((....(((.((....)).)))...)))...))............ ( -21.80, z-score = -2.99, R) >droYak2.chr2R 4594866 91 + 21139217 ----GCAUAUGUUUUCCAUUA-AAGUGGAAAAUAAAUAUGCUACCGG-AAAACUAGAUAUGAGUGAAAACACAUUGAAGUGUGCCCCAUCUGCAGAC ----(((((((((((((((..-..)))))))))..))))))....((-...(((....(((.((....)).)))...)))...))............ ( -21.80, z-score = -2.99, R) >droSim1.chr2R 11174960 91 - 19596830 ----GCAUAUGUUUUCCAUUA-AAGUGGAAAAUAAAUAUGCUACCGG-AAAACUAGAUAUGAGUGAAAACACAUUGAAGUGUGCCCCAUCUGCAGAC ----(((((((((((((((..-..)))))))))..))))))....((-...(((....(((.((....)).)))...)))...))............ ( -21.80, z-score = -2.99, R) >droSec1.super_1 9939455 91 - 14215200 ----GCAUAUGUUUUCCAUUA-AAGUGGAAAAUAAAUAUGCUACCGG-AAAACUAGAUAUGAGUGAAAACACAUUGAAGUGUGCCCCAUCUGCACAC ----(((((((((((((((..-..)))))))))..))))))......-..........(((.((....)).)))....((((((.......)))))) ( -23.90, z-score = -3.59, R) >consensus ____GCAUAUGUUUUCCAUUA_AAGUGGAAAAUAAAUAUGCUACCGG_AAAACUAGAUAUGAGUGAAAACACAUUGAAGUGUGCCCCAUCUGCAGAC ....(((((((((((((((.....))))))))))..)))))....((....((.....(((.((....)).)))......))...)).......... (-19.86 = -19.43 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:27:39 2011