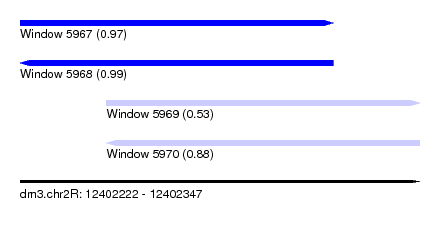
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,402,222 – 12,402,347 |
| Length | 125 |
| Max. P | 0.991836 |

| Location | 12,402,222 – 12,402,320 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.99 |
| Shannon entropy | 0.40227 |
| G+C content | 0.37078 |
| Mean single sequence MFE | -27.21 |
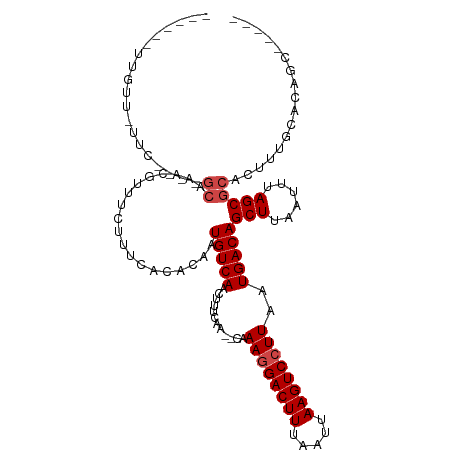
| Consensus MFE | -16.07 |
| Energy contribution | -16.76 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
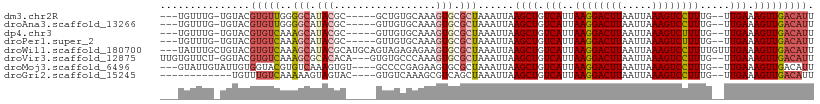
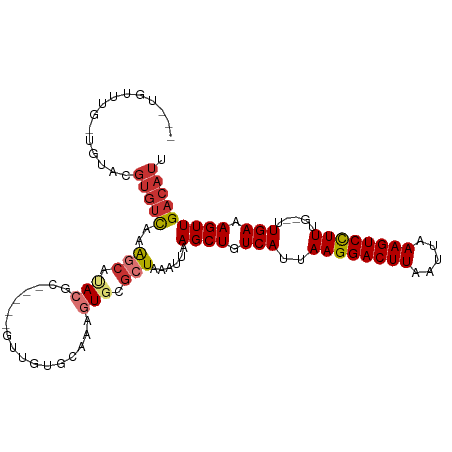
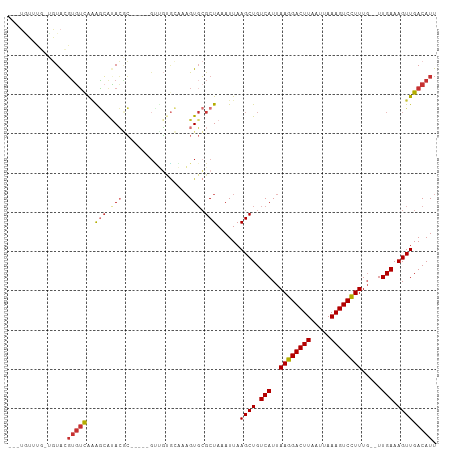
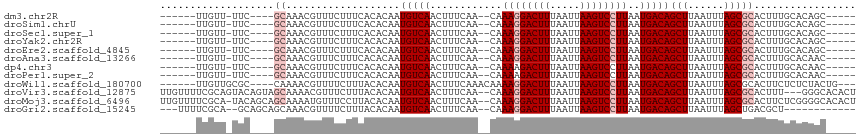
>dm3.chr2R 12402222 98 + 21146708 ---UGUUUG-UGUACGUGUUGGGGCAUACGC-----GCUGUGCAAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUUG--UUGAAAGUUGACAUU ---((((.(-(((((.((((((.((....))-----.))).)))..)))))).......((((.(((..((((((((.....))))))))..--.))).)))))))).. ( -28.30, z-score = -1.81, R) >droAna3.scaffold_13266 15776083 98 + 19884421 ---UGUUUG-UGUACGUGUUGGGGCAUACGC-----GUUGUGCAAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUUG--UUGAAAGUUGACAUU ---.....(-(((((((((........))))-----)).((((.....)))).......((((.(((..((((((((.....))))))))..--.))).)))).)))). ( -26.50, z-score = -1.51, R) >dp4.chr3 16040017 98 - 19779522 ---UGUUUG-UGUACGUGUCAAAGCAUACGC-----GUUGUGCAAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCUUUUG--UUGAAAGUUGACAUU ---((((.(-(((((.((((((.((....))-----.))).)))..)))))).......((((.(((..((((((((.....))))))))..--.))).)))))))).. ( -25.00, z-score = -1.31, R) >droPer1.super_2 5109650 98 - 9036312 ---UGUUUG-UGUACGUGUCAAAGCAUACGC-----GUUGUGCAAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCUUUUG--UUGAAAGUUGACAUU ---((((.(-(((((.((((((.((....))-----.))).)))..)))))).......((((.(((..((((((((.....))))))))..--.))).)))))))).. ( -25.00, z-score = -1.31, R) >droWil1.scaffold_180700 857978 106 + 6630534 ---UAUUUGCUGUACGUGUCAAAGCAUACGCAUGCAGUAGAGAGAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUUUGUUUGAAAGUUGACAUU ---..(((((((((.((((........)))).))))))))).........(((......)))(((((..((((((((.....))))))))............))))).. ( -27.14, z-score = -1.57, R) >droVir3.scaffold_12875 18926843 103 + 20611582 UUGUGUUCU-GGUACGUGUCAAAGCGCACACA---GUGUGCCCAAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUUG--UUGAAAGUUGACAUU .........-.....(((((..(((((((...---.((....))..)))))))......((((.(((..((((((((.....))))))))..--.))).))))))))). ( -31.10, z-score = -2.83, R) >droMoj3.scaffold_6496 12611279 100 + 26866924 ---GUAUUGUAUUGUGGUACGUGUCAAAGUGU----GCCCCGAGAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUUG--UUGAAAGUUGACAUU ---((((..(...)..))))(((((..((((.----.(........)..))))......((((.(((..((((((((.....))))))))..--.))).))))))))). ( -29.80, z-score = -4.22, R) >droGri2.scaffold_15245 5003512 91 + 18325388 ------------UGUUUGUCAAAAAGUAGUAC----GUGUCAAAGCGUCAGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUUG--UUGAAAGUUGACAUU ------------....((((((........((----((......))))(((((......)))))(((..((((((((.....))))))))..--.)))...)))))).. ( -24.80, z-score = -3.97, R) >consensus ___UGUUUG_UGUACGUGUCAAAGCAUACGC_____GUUGUGCAAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUUG__UUGAAAGUUGACAUU ...............(((((..(((.(((.................))).)))......((((.(((..((((((((.....)))))))).....))).))))))))). (-16.07 = -16.76 + 0.69)
| Location | 12,402,222 – 12,402,320 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.99 |
| Shannon entropy | 0.40227 |
| G+C content | 0.37078 |
| Mean single sequence MFE | -21.00 |
| Consensus MFE | -14.12 |
| Energy contribution | -14.51 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.991836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12402222 98 - 21146708 AAUGUCAACUUUCAA--CAAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUUGCACAGC-----GCGUAUGCCCCAACACGUACA-CAAACA--- ..(((((........--..((((((((.....))))))))..)))))..........((((..........))-----))(((((........))))).-......--- ( -21.30, z-score = -2.64, R) >droAna3.scaffold_13266 15776083 98 - 19884421 AAUGUCAACUUUCAA--CAAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUUGCACAAC-----GCGUAUGCCCCAACACGUACA-CAAACA--- ..(((((........--..((((((((.....))))))))..)))))(((......)))((.....)).....-----(.(((((........))))).-).....--- ( -20.00, z-score = -2.72, R) >dp4.chr3 16040017 98 + 19779522 AAUGUCAACUUUCAA--CAAAAGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUUGCACAAC-----GCGUAUGCUUUGACACGUACA-CAAACA--- ..((((((.......--.....(((((.....)))))..........(((......)))(((...(((.....-----)))..))).))))))......-......--- ( -16.50, z-score = -0.89, R) >droPer1.super_2 5109650 98 + 9036312 AAUGUCAACUUUCAA--CAAAAGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUUGCACAAC-----GCGUAUGCUUUGACACGUACA-CAAACA--- ..((((((.......--.....(((((.....)))))..........(((......)))(((...(((.....-----)))..))).))))))......-......--- ( -16.50, z-score = -0.89, R) >droWil1.scaffold_180700 857978 106 - 6630534 AAUGUCAACUUUCAAACAAAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUCUCUCUACUGCAUGCGUAUGCUUUGACACGUACAGCAAAUA--- ..(((((............((((((((.....))))))))..)))))(((......)))(((...........))).((((((((........))))).)))....--- ( -21.64, z-score = -1.78, R) >droVir3.scaffold_12875 18926843 103 - 20611582 AAUGUCAACUUUCAA--CAAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUUGGGCACAC---UGUGUGCGCUUUGACACGUACC-AGAACACAA ..((((((((.(((.--..((((((((.....))))))))..))).))........(((((((..(((.....)---)).)))))))))))))......-......... ( -28.80, z-score = -3.24, R) >droMoj3.scaffold_6496 12611279 100 - 26866924 AAUGUCAACUUUCAA--CAAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUCUCGGGGC----ACACUUUGACACGUACCACAAUACAAUAC--- ..((((((...(((.--..((((((((.....))))))))..)))..(((......)))((.((.....))))----.....)))))).(((......))).....--- ( -20.30, z-score = -3.20, R) >droGri2.scaffold_15245 5003512 91 - 18325388 AAUGUCAACUUUCAA--CAAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCUGACGCUUUGACAC----GUACUACUUUUUGACAAACA------------ ..((((((...(((.--..((((((((.....))))))))..)))(((((......)))))(((........)----))........))))))....------------ ( -23.00, z-score = -4.71, R) >consensus AAUGUCAACUUUCAA__CAAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUUGCACAAC_____GCGUAUGCUUUGACACGUACA_CAAACA___ ..(((((............((((((((.....))))))))..)))))(((......))).....................(((((........)))))........... (-14.12 = -14.51 + 0.39)

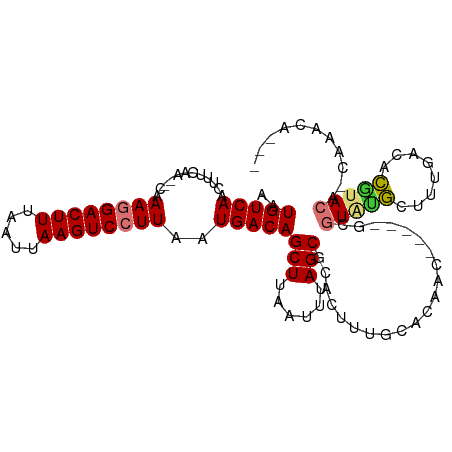
| Location | 12,402,249 – 12,402,347 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 84.89 |
| Shannon entropy | 0.30022 |
| G+C content | 0.35478 |
| Mean single sequence MFE | -23.87 |
| Consensus MFE | -17.29 |
| Energy contribution | -17.09 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.526411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
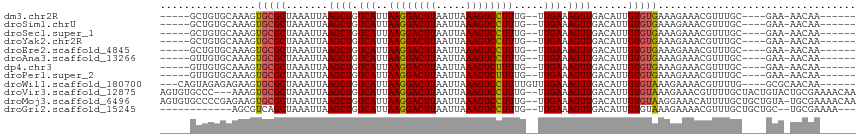
>dm3.chr2R 12402249 98 + 21146708 -----GCUGUGCAAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUUG--UUGAAAGUUGACAUUGUGUGAAAGAAACGUUUGC----GAA-AACAA------ -----....((((((((((((.......((((.(((..((((((((.....))))))))..--.))).))))......)))).......)).)))))----)..-.....------ ( -22.83, z-score = -1.29, R) >droSim1.chrU 11210441 98 + 15797150 -----GCUGUGCAAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUUG--UUGAAAGUUGACAUUGUGUGAAAGAAACGUUUGC----GAA-AACAA------ -----....((((((((((((.......((((.(((..((((((((.....))))))))..--.))).))))......)))).......)).)))))----)..-.....------ ( -22.83, z-score = -1.29, R) >droSec1.super_1 9906139 98 + 14215200 -----GCUGUGCAAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUUG--UUGAAAGUUGACAUUGUGUGAAAGAAACGUUUGC----GAA-AACAA------ -----....((((((((((((.......((((.(((..((((((((.....))))))))..--.))).))))......)))).......)).)))))----)..-.....------ ( -22.83, z-score = -1.29, R) >droYak2.chr2R 4559393 98 - 21139217 -----GCUGUGCAAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUUG--UUGAAAGUUGACAUUGUGUGAAAGAAACGUUUGC----GAA-AACAA------ -----....((((((((((((.......((((.(((..((((((((.....))))))))..--.))).))))......)))).......)).)))))----)..-.....------ ( -22.83, z-score = -1.29, R) >droEre2.scaffold_4845 9134971 98 - 22589142 -----GCUGUGCAAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUUG--UUGAAAGUUGACAUUGUGUGAAAGAAACGUUUGC----GAA-AACAA------ -----....((((((((((((.......((((.(((..((((((((.....))))))))..--.))).))))......)))).......)).)))))----)..-.....------ ( -22.83, z-score = -1.29, R) >droAna3.scaffold_13266 15776110 98 + 19884421 -----GUUGUGCAAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUUG--UUGAAAGUUGACAUUGUGUGAAAGAAACGUUUGC----GAA-AACAA------ -----(((.((((((((((((.......((((.(((..((((((((.....))))))))..--.))).))))......)))).......)).)))))----)..-)))..------ ( -23.93, z-score = -1.90, R) >dp4.chr3 16040044 98 - 19779522 -----GUUGUGCAAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCUUUUG--UUGAAAGUUGACAUUGUGUGAAAGAAACGUUUGC----GAA-AACAA------ -----(((.((((((((((((.......((((.(((..((((((((.....))))))))..--.))).))))......)))).......)).)))))----)..-)))..------ ( -21.23, z-score = -0.72, R) >droPer1.super_2 5109677 98 - 9036312 -----GUUGUGCAAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCUUUUG--UUGAAAGUUGACAUUGUGUGAAAGAAACGUUUGC----GAA-AACAA------ -----(((.((((((((((((.......((((.(((..((((((((.....))))))))..--.))).))))......)))).......)).)))))----)..-)))..------ ( -21.23, z-score = -0.72, R) >droWil1.scaffold_180700 858009 103 + 6630534 ---CAGUAGAGAGAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUUUGUUUGAAAGUUGACAUUGUGUAAAGAAAACGUUUUG----GCGCAACAA------ ---.............(((((((((...((((.(((..((((((((.....)))))))).....))).)))).((.....))...........))))----)))))....------ ( -25.00, z-score = -1.74, R) >droVir3.scaffold_12875 18926873 111 + 20611582 AGUGUGCCC---AAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUUG--UUGAAAGUUGACAUUGUGUAAAGAAACGUUUUGCUACUGUACUGCGAAAACAA ((((..(..---...)..))))......((((.(((..((((((((.....))))))))..--.))).)))).................(((..(((....)))..)))....... ( -26.30, z-score = -1.63, R) >droMoj3.scaffold_6496 12611303 113 + 26866924 AGUGUGCCCCGAGAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUUG--UUGAAAGUUGACAUUGUGUAAGGAAACAUUUUGCUGCUGUA-UGCGAAAACAA ((((..(........)..))))......((((.(((..((((((((.....))))))))..--.))).))))....((((....(....).((((((.......-.)))))))))) ( -29.00, z-score = -2.11, R) >droGri2.scaffold_15245 5003539 97 + 18325388 ------------AGCGUCAGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUUG--UUGAAAGUUGACAUUGUGUAAAGAAAACGUUUGCUGCUGC--UGCGAAAA--- ------------.(((.((((.......((((.(((..((((((((.....))))))))..--.))).))))........(((((.......))))).)))).--))).....--- ( -25.60, z-score = -2.11, R) >consensus _____GCUGUGCAAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUUG__UUGAAAGUUGACAUUGUGUGAAAGAAACGUUUGC____GAA_AACAA______ ................(((((.......((((.(((..((((((((.....)))))))).....))).))))......)))))................................. (-17.29 = -17.09 + -0.19)

| Location | 12,402,249 – 12,402,347 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 84.89 |
| Shannon entropy | 0.30022 |
| G+C content | 0.35478 |
| Mean single sequence MFE | -18.77 |
| Consensus MFE | -13.78 |
| Energy contribution | -14.28 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12402249 98 - 21146708 ------UUGUU-UUC----GCAAACGUUUCUUUCACACAAUGUCAACUUUCAA--CAAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUUGCACAGC----- ------.....-...----(((((.((.............(((((........--..((((((((.....))))))))..)))))(((......)))..))))))).....----- ( -19.30, z-score = -2.25, R) >droSim1.chrU 11210441 98 - 15797150 ------UUGUU-UUC----GCAAACGUUUCUUUCACACAAUGUCAACUUUCAA--CAAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUUGCACAGC----- ------.....-...----(((((.((.............(((((........--..((((((((.....))))))))..)))))(((......)))..))))))).....----- ( -19.30, z-score = -2.25, R) >droSec1.super_1 9906139 98 - 14215200 ------UUGUU-UUC----GCAAACGUUUCUUUCACACAAUGUCAACUUUCAA--CAAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUUGCACAGC----- ------.....-...----(((((.((.............(((((........--..((((((((.....))))))))..)))))(((......)))..))))))).....----- ( -19.30, z-score = -2.25, R) >droYak2.chr2R 4559393 98 + 21139217 ------UUGUU-UUC----GCAAACGUUUCUUUCACACAAUGUCAACUUUCAA--CAAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUUGCACAGC----- ------.....-...----(((((.((.............(((((........--..((((((((.....))))))))..)))))(((......)))..))))))).....----- ( -19.30, z-score = -2.25, R) >droEre2.scaffold_4845 9134971 98 + 22589142 ------UUGUU-UUC----GCAAACGUUUCUUUCACACAAUGUCAACUUUCAA--CAAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUUGCACAGC----- ------.....-...----(((((.((.............(((((........--..((((((((.....))))))))..)))))(((......)))..))))))).....----- ( -19.30, z-score = -2.25, R) >droAna3.scaffold_13266 15776110 98 - 19884421 ------UUGUU-UUC----GCAAACGUUUCUUUCACACAAUGUCAACUUUCAA--CAAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUUGCACAAC----- ------.....-...----(((((.((.............(((((........--..((((((((.....))))))))..)))))(((......)))..))))))).....----- ( -19.30, z-score = -2.78, R) >dp4.chr3 16040044 98 + 19779522 ------UUGUU-UUC----GCAAACGUUUCUUUCACACAAUGUCAACUUUCAA--CAAAAGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUUGCACAAC----- ------.....-...----(((((.((.............(((((..(((...--..)))(((((.....))))).....)))))(((......)))..))))))).....----- ( -13.90, z-score = -0.92, R) >droPer1.super_2 5109677 98 + 9036312 ------UUGUU-UUC----GCAAACGUUUCUUUCACACAAUGUCAACUUUCAA--CAAAAGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUUGCACAAC----- ------.....-...----(((((.((.............(((((..(((...--..)))(((((.....))))).....)))))(((......)))..))))))).....----- ( -13.90, z-score = -0.92, R) >droWil1.scaffold_180700 858009 103 - 6630534 ------UUGUUGCGC----CAAAACGUUUUCUUUACACAAUGUCAACUUUCAAACAAAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUCUCUCUACUG--- ------....(((((----.(((..((.......))....(((((............((((((((.....))))))))..)))))......))).))))).............--- ( -18.54, z-score = -2.33, R) >droVir3.scaffold_12875 18926873 111 - 20611582 UUGUUUUCGCAGUACAGUAGCAAAACGUUUCUUUACACAAUGUCAACUUUCAA--CAAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUU---GGGCACACU ........((.(((.((.(((.....))).)).))).(((.(((.....(((.--..((((((((.....))))))))..)))..(((......)))).)).))---).))..... ( -21.00, z-score = -1.19, R) >droMoj3.scaffold_6496 12611303 113 - 26866924 UUGUUUUCGCA-UACAGCAGCAAAAUGUUUCCUUACACAAUGUCAACUUUCAA--CAAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUCUCGGGGCACACU (((((...((.-....)))))))..(((.((((.......(((((........--..((((((((.....))))))))..)))))(((......))).........)))).))).. ( -21.30, z-score = -1.45, R) >droGri2.scaffold_15245 5003539 97 - 18325388 ---UUUUCGCA--GCAGCAGCAAACGUUUUCUUUACACAAUGUCAACUUUCAA--CAAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCUGACGCU------------ ---.....((.--...))(((............................(((.--..((((((((.....))))))))..)))(((((......)))))..)))------------ ( -20.80, z-score = -2.34, R) >consensus ______UUGUU_UUC____GCAAACGUUUCUUUCACACAAUGUCAACUUUCAA__CAAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUUGCACAGC_____ ...................((...................(((((............((((((((.....))))))))..)))))(((......)))))................. (-13.78 = -14.28 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:27:35 2011