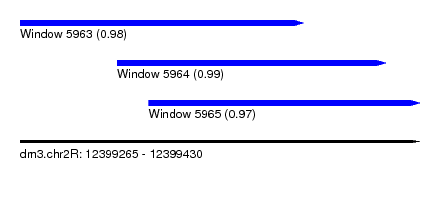
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,399,265 – 12,399,430 |
| Length | 165 |
| Max. P | 0.986472 |

| Location | 12,399,265 – 12,399,382 |
|---|---|
| Length | 117 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.77 |
| Shannon entropy | 0.29253 |
| G+C content | 0.50819 |
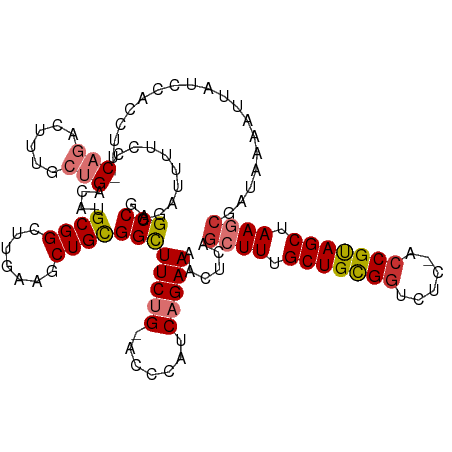
| Mean single sequence MFE | -38.52 |
| Consensus MFE | -25.70 |
| Energy contribution | -26.09 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
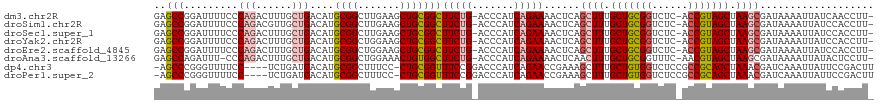
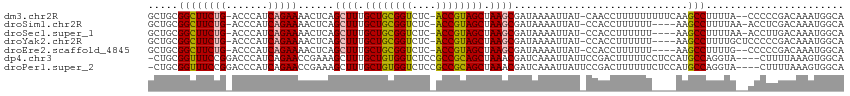
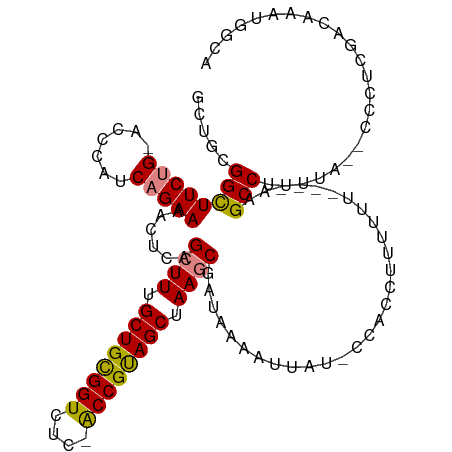
>dm3.chr2R 12399265 117 + 21146708 GAGCCGGAUUUUCCCAGACUUUGCUGACAUGCGGCUUGAAGCUGCGGCUUCUG-ACCCAUCAGAAAACUCAGCUUUGCUGCGGUCUC-ACCGUAGCUAAGCGAUAAAAUUAUCAACCUU- (((((((......)).......((......)))))))(((((((.(..(((((-(....))))))..).)))))))(((((((....-.))))))).....((((....))))......- ( -41.20, z-score = -3.66, R) >droSim1.chr2R 11138409 117 + 19596830 GAGCCGGAUUUUCCCAGACGUUGCUGACAUGCGGCUUGAAGCUGCGGCUUCUG-ACCCAUCAGAAAACUCAGCUUUGCUGCGGUCUC-ACCGUAGCUAAGCGAUAAAAUUAUCCACCUU- (((((((......)).......((......)))))))(((((((.(..(((((-(....))))))..).)))))))(((((((....-.)))))))...(.((((....))))).....- ( -40.60, z-score = -3.16, R) >droSec1.super_1 9903117 117 + 14215200 GAGCCGGAUUUUCCCAGACUUUGCUGACAUGCGGCUUGAAGCUGCGGCUUCUG-ACCCAUCAGAAAACUCAGCUUUGCUGCGGUCUC-ACCGUAGCUAAGCGAUAAAAUUAUCCACCUU- (((((((......)).......((......)))))))(((((((.(..(((((-(....))))))..).)))))))(((((((....-.)))))))...(.((((....))))).....- ( -40.60, z-score = -3.48, R) >droYak2.chr2R 4556571 117 - 21139217 GAGCCGGAUUUUCCCAGACUUUGCUGACAUGCGGCUGGAAGCUGCGGCUUCUG-ACCCAUCAGAAAACUCAGCUUUGCUGCGGUCUC-ACCGUAGCUAAGCGAUAAAAUUAUCCACCUU- .....((((.......((.(((.((((..((.((..((((((....)))))).-.)))))))).))).)).((((.(((((((....-.))))))).)))).........)))).....- ( -40.00, z-score = -2.80, R) >droEre2.scaffold_4845 9132209 117 - 22589142 GAGCCGGAUUUUCCCAGACUUUGCUGACAUGCGGCUGGAAGCUGCGGCUUCUG-ACCCAUCAGAAAACUCAGCUUUGCUGCGGUCUC-ACCGUAGCUAAGCGAUAAAAUUAUCCACCUU- .....((((.......((.(((.((((..((.((..((((((....)))))).-.)))))))).))).)).((((.(((((((....-.))))))).)))).........)))).....- ( -40.00, z-score = -2.80, R) >droAna3.scaffold_13266 15773588 116 + 19884421 GAGCCAGAUUU-CCCAGACUUUGCUGACAUGCGGCUGGAAACUGUGGCUUCUG-ACCCAUCAGAAAACUCAACUUUGCUGCGGUUUC-AACGUAGCUAAGCGAUAAAAUUAUACUCCUU- ..((((.(.((-.((((....(((......))).)))).)).).))))(((((-(....)))))).......(((.((((((.....-..)))))).)))...................- ( -30.20, z-score = -1.57, R) >dp4.chr3 16037557 114 - 19779522 -AGCCCGGGUUUUCC----UCUGAUGACAUGCGGCUUUCC-CUGCGGUUUCCGGACCCAUCAGAACCGAAAGCUUUGCUGUGGUCUCCGCCGCAGCUAAACGAUCAAAUUAUUCCGACUU -....(((((((((.----(((((((....((((......-))))((((....)))))))))))...))))))...((((((((....)))))))).................))).... ( -37.80, z-score = -2.38, R) >droPer1.super_2 5107285 114 - 9036312 -AGCCCGGGUUUUCC----UCUGAUGACAUGCGGCUUUCC-CUGCGGUUUCCGGACCCAUCAGAACCGAAAGCUUUGCUGUGGUCUCCGCCGCAGCUAAACGAUCAAAUUAUUCCGACUU -....(((((((((.----(((((((....((((......-))))((((....)))))))))))...))))))...((((((((....)))))))).................))).... ( -37.80, z-score = -2.38, R) >consensus GAGCCGGAUUUUCCCAGACUUUGCUGACAUGCGGCUUGAAGCUGCGGCUUCUG_ACCCAUCAGAAAACUCAGCUUUGCUGCGGUCUC_ACCGUAGCUAAGCGAUAAAAUUAUCCACCUU_ (((((.........(((......)))....((((.......))))))))).....................((((.((((((((....)))))))).))))................... (-25.70 = -26.09 + 0.39)
| Location | 12,399,305 – 12,399,416 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 82.87 |
| Shannon entropy | 0.30669 |
| G+C content | 0.47312 |
| Mean single sequence MFE | -30.24 |
| Consensus MFE | -22.83 |
| Energy contribution | -22.59 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.986472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
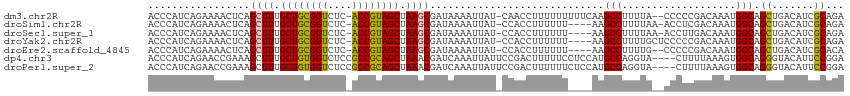
>dm3.chr2R 12399305 111 + 21146708 GCUGCGGCUUCUG-ACCCAUCAGAAAACUCAGCUUUGCUGCGGUCUC-ACCGUAGCUAAGCGAUAAAAUUAU-CAACCUUUUUUUUUCAAGCCUUUUA--CCCCCGACAAAUGGCA ((((.(..(((((-(....))))))..).))))...(((((((....-.))))))).....((((....)))-)................(((.....--............))). ( -30.43, z-score = -3.29, R) >droSim1.chr2R 11138449 108 + 19596830 GCUGCGGCUUCUG-ACCCAUCAGAAAACUCAGCUUUGCUGCGGUCUC-ACCGUAGCUAAGCGAUAAAAUUAU-CCACCUUUUUU----AAGCCUUUUAA-ACCUCGACAAAUGGCA ((((.(..(((((-(....))))))..).))))...(((((((....-.)))))))...(.((((....)))-)).........----..(((......-............))). ( -29.77, z-score = -3.04, R) >droSec1.super_1 9903157 108 + 14215200 GCUGCGGCUUCUG-ACCCAUCAGAAAACUCAGCUUUGCUGCGGUCUC-ACCGUAGCUAAGCGAUAAAAUUAU-CCACCUUUUUU----AAGCCUUUUAA-ACCUUGACAAAUGGCA ((((.(..(((((-(....))))))..).))))...(((((((....-.)))))))...(.((((....)))-)).........----..(((......-............))). ( -29.77, z-score = -2.98, R) >droYak2.chr2R 4556611 109 - 21139217 GCUGCGGCUUCUG-ACCCAUCAGAAAACUCAGCUUUGCUGCGGUCUC-ACCGUAGCUAAGCGAUAAAAUUAU-CCACCUUUUUU----AAGCCUUUUGCUCCCCCGACAAAUGGCA ((((.(..(((((-(....))))))..).))))...(((((((....-.)))))))...(.((((....)))-)).........----..(((.((((.((....)))))).))). ( -33.80, z-score = -3.87, R) >droEre2.scaffold_4845 9132249 107 - 22589142 GCUGCGGCUUCUG-ACCCAUCAGAAAACUCAGCUUUGCUGCGGUCUC-ACCGUAGCUAAGCGAUAAAAUUAU-CCACCUUUUUU----AAGCCUUUUG--CCCCCGACAAAUGGCA ((((.(..(((((-(....))))))..).))))...(((((((....-.)))))))...(.((((....)))-)).........----..(((.((((--.......)))).))). ( -32.90, z-score = -3.62, R) >dp4.chr3 16037592 111 - 19779522 -CUGCGGUUUCCGGACCCAUCAGAACCGAAAGCUUUGCUGUGGUCUCCGCCGCAGCUAAACGAUCAAAUUAUUCCGACUUUUUCCUCCAUGCCAGGUA----CUUUUAAAGUGGCA -(((((((....(((((((.((((..(....).)))).)).)))))..)))))))..................((.(((((.....((......))..----.....))))))).. ( -27.50, z-score = -0.57, R) >droPer1.super_2 5107320 111 - 9036312 -CUGCGGUUUCCGGACCCAUCAGAACCGAAAGCUUUGCUGUGGUCUCCGCCGCAGCUAAACGAUCAAAUUAUUCCGACUUUUUUCUCCAUGCCAGGUA----CUUUUAAAGUGGCA -(((((((....(((((((.((((..(....).)))).)).)))))..)))))))..................((.(((((.....((......))..----.....))))))).. ( -27.50, z-score = -0.59, R) >consensus GCUGCGGCUUCUG_ACCCAUCAGAAAACUCAGCUUUGCUGCGGUCUC_ACCGUAGCUAAGCGAUAAAAUUAU_CCACCUUUUUU____AAGCCUUUUA__CCCUCGACAAAUGGCA .....((((((((.......)))))......((((.((((((((....)))))))).)))).............................)))....................... (-22.83 = -22.59 + -0.24)
| Location | 12,399,318 – 12,399,430 |
|---|---|
| Length | 112 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.85 |
| Shannon entropy | 0.32667 |
| G+C content | 0.45976 |
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -21.81 |
| Energy contribution | -21.08 |
| Covariance contribution | -0.73 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12399318 112 + 21146708 ACCCAUCAGAAAACUCAGCUUUGCUGCGGUCUC-ACCGUAGCUAAGCGAUAAAAUUAU-CAACCUUUUUUUUUCAAGCCUUUUA--CCCCCGACAAAUGGCAGCUGACAUCGCAGA ........(((((....((((.(((((((....-.))))))).))))((((....)))-).........)))))..(((.....--............)))..(((......))). ( -27.23, z-score = -2.85, R) >droSim1.chr2R 11138462 109 + 19596830 ACCCAUCAGAAAACUCAGCUUUGCUGCGGUCUC-ACCGUAGCUAAGCGAUAAAAUUAU-CCACCUUUUUU----AAGCCUUUUAA-ACCUCGACAAAUGGCAGCUGACAUCGCAGA .....((((........((((.(((((((....-.))))))).))))((((....)))-)..........----..(((......-............)))..))))......... ( -25.47, z-score = -2.13, R) >droSec1.super_1 9903170 109 + 14215200 ACCCAUCAGAAAACUCAGCUUUGCUGCGGUCUC-ACCGUAGCUAAGCGAUAAAAUUAU-CCACCUUUUUU----AAGCCUUUUAA-ACCUUGACAAAUGGCAGCUGACAUCGCAGA .....((((........((((.(((((((....-.))))))).))))((((....)))-)..........----..(((......-............)))..))))......... ( -25.47, z-score = -2.04, R) >droYak2.chr2R 4556624 110 - 21139217 ACCCAUCAGAAAACUCAGCUUUGCUGCGGUCUC-ACCGUAGCUAAGCGAUAAAAUUAU-CCACCUUUUUU----AAGCCUUUUGCUCCCCCGACAAAUGGCAGCUGACAUCGCAGA .....((((........((((.(((((((....-.))))))).))))((((....)))-)..........----..(((.((((.((....)))))).)))..))))......... ( -29.50, z-score = -3.13, R) >droEre2.scaffold_4845 9132262 108 - 22589142 ACCCAUCAGAAAACUCAGCUUUGCUGCGGUCUC-ACCGUAGCUAAGCGAUAAAAUUAU-CCACCUUUUUU----AAGCCUUUUG--CCCCCGACAAAUGGCAGCUGACAUCGCACA .....((((........((((.(((((((....-.))))))).))))((((....)))-)..........----..(((.((((--.......)))).)))..))))......... ( -28.60, z-score = -3.28, R) >dp4.chr3 16037605 112 - 19779522 ACCCAUCAGAACCGAAAGCUUUGCUGUGGUCUCCGCCGCAGCUAAACGAUCAAAUUAUUCCGACUUUUUCCUCCAUGCCAGGUA----CUUUUAAAGUGGCAGGGUACAUUCCGGA (((((((.(((.(....).)))((((((((....)))))))).....))).........................(((((..(.----......)..))))))))).......... ( -26.60, z-score = -0.25, R) >droPer1.super_2 5107333 112 - 9036312 ACCCAUCAGAACCGAAAGCUUUGCUGUGGUCUCCGCCGCAGCUAAACGAUCAAAUUAUUCCGACUUUUUUCUCCAUGCCAGGUA----CUUUUAAAGUGGCAGGGUACAUUCCGGA (((((((.(((.(....).)))((((((((....)))))))).....))).........................(((((..(.----......)..))))))))).......... ( -26.60, z-score = -0.31, R) >consensus ACCCAUCAGAAAACUCAGCUUUGCUGCGGUCUC_ACCGUAGCUAAGCGAUAAAAUUAU_CCACCUUUUUU____AAGCCUUUUA__CCCUCGACAAAUGGCAGCUGACAUCGCAGA .................((((.((((((((....)))))))).)))).............................(((...................))).((.......))... (-21.81 = -21.08 + -0.73)
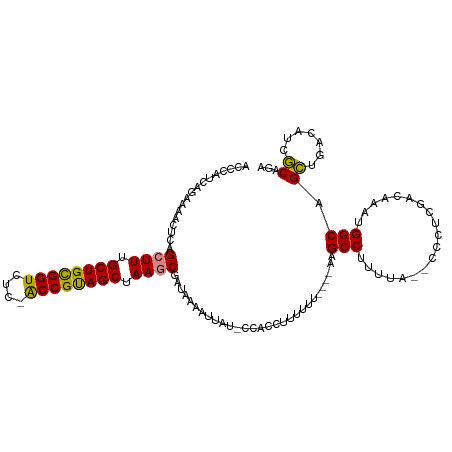
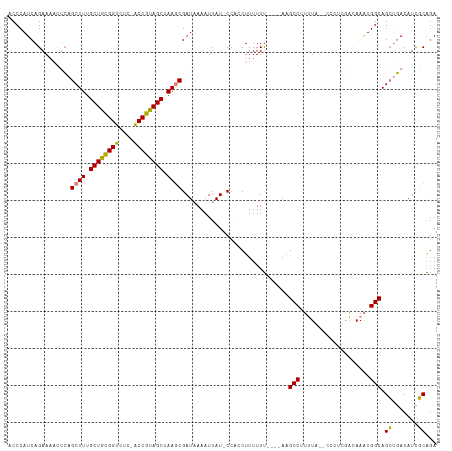
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:27:31 2011