
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,397,336 – 12,397,435 |
| Length | 99 |
| Max. P | 0.558596 |

| Location | 12,397,336 – 12,397,435 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 69.75 |
| Shannon entropy | 0.54898 |
| G+C content | 0.47509 |
| Mean single sequence MFE | -20.34 |
| Consensus MFE | -10.19 |
| Energy contribution | -10.97 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.558596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
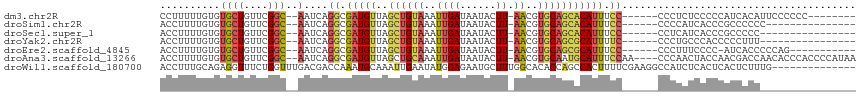
>dm3.chr2R 12397336 99 + 21146708 CCUUUUUGUGUGCUGUUCGGC--AAUCAGGCGAUGUUAGCUGUAAAUUGAUAAUACUU-AACGUGCAGCACAUUUCC------CCCUCUCCCCCAUCACAUUCCCCCC-------- ......((((((((....)))--)....((.(((((..((((((..((((......))-))..))))))))))).))------.............))))........-------- ( -21.00, z-score = -2.36, R) >droSim1.chr2R 11136477 92 + 19596830 ACCUUUUGUGUGCUGUUCGGC--AAUCAGGCGAUGUUAGCUGUAAAUUGAUAAUACUU-AACGUGCAGCACAUUUCC------CCCCAUCACCCGCCCCCC--------------- .......(((((((....)))--)....((.(((((..((((((..((((......))-))..))))))))))).))------......))).........--------------- ( -19.40, z-score = -1.11, R) >droSec1.super_1 9901174 91 + 14215200 ACCUUUUGUGUGCUGUUCGGC--AAUCAGGCGAUGUUAGCUGUAAAUUGAUAAUACUU-AACGUGCAGCACAUUUCC------CCUCAUCACCCGCCCCC---------------- .......(((((((....)))--)....((.(((((..((((((..((((......))-))..))))))))))).))------......)))........---------------- ( -19.40, z-score = -1.01, R) >droYak2.chr2R 4554601 91 - 21139217 ACCUUUUGUGUGCUGUUCGGC--AAUCAGGCGAUGUUAGCUGUAAAUUGAUAAUACUU-AACGUGCAGCGCAUUUUC------CCCUGCCCACCCCCUUU---------------- .......(((((((....)))--)..((((.((.....((((((..((((......))-))..))))))......))------.))))..))).......---------------- ( -20.90, z-score = -1.34, R) >droEre2.scaffold_4845 9130352 95 - 22589142 ACCUUUUGUGUGCUGUUCGGC--AAUCAGGCGAUGUUAGCUGUAAAUUGAUAAUACUU-AACGUGCAGCGCAUUUCC------CCCUUUCCCC-AUCACCCCCAG----------- .......(((((((....)))--)....((.(((((..((((((..((((......))-))..))))))))))).))------..........-..)))......----------- ( -19.70, z-score = -1.38, R) >droAna3.scaffold_13266 15771584 109 + 19884421 ACCUUUUGUGUGCUGUUCGGC--AAUCAGGCGAUGUUAGCUGCAAAUUGAUAAUACUU-AACGUGCAAUGCAUUUCCAA----CCCAACUACCAACGACCAACACCCACCCCAUAA .......(((((((....)))--.....((.(((((....((((..((((......))-))..))))..))))).))..----..................))))........... ( -17.80, z-score = 0.32, R) >droWil1.scaffold_180700 5783787 102 - 6630534 ACCUUUGCAGAGGUUUCUGGUUUGACGACCAAAUGCAAAUUGAAUAUGGAGAAUGCUUUGGCACACCAGCCACUUUUCGAAGGCCAUCUCACUCACUCUUUG-------------- .((((((.((((((..(((((.((....(((((.(((................)))))))))).)))))..))))))))))))...................-------------- ( -24.19, z-score = -0.70, R) >consensus ACCUUUUGUGUGCUGUUCGGC__AAUCAGGCGAUGUUAGCUGUAAAUUGAUAAUACUU_AACGUGCAGCACAUUUCC______CCCUAUCACCCACCACC________________ .......(((((((((.((...........((((...........))))............)).)))))))))........................................... (-10.19 = -10.97 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:27:29 2011