| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,048,685 – 3,048,844 |
| Length | 159 |
| Max. P | 0.994842 |

| Location | 3,048,685 – 3,048,844 |
|---|---|
| Length | 159 |
| Sequences | 12 |
| Columns | 161 |
| Reading direction | forward |
| Mean pairwise identity | 79.95 |
| Shannon entropy | 0.44051 |
| G+C content | 0.40226 |
| Mean single sequence MFE | -45.98 |
| Consensus MFE | -28.17 |
| Energy contribution | -27.07 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.994842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
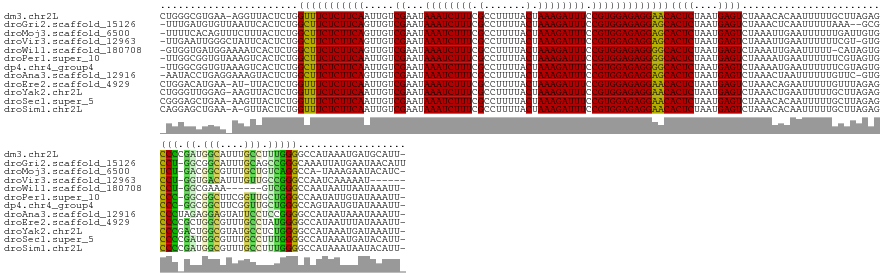
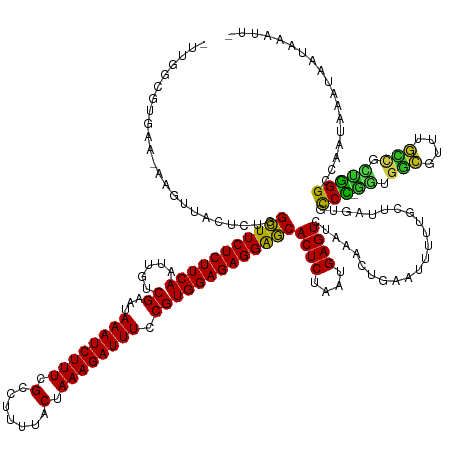
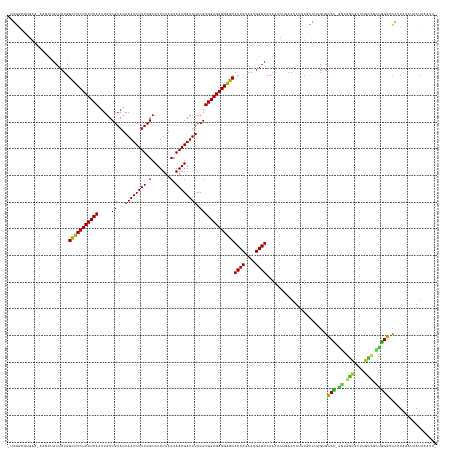
>dm3.chr2L 3048685 159 + 23011544 CUGGGCGUGAA-AGGUUACUCUGGUUUCUCUUCAAUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUAAACACAAUUUUUGCUUAGAGCCCCGAUGGCAUUUGCCUUUGGGGCCAUAAAUGAUGCAUU- (((((((.(((-(.(((......(((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....))))...)))....))))))))))).(((((((.(((....))).)))))))...............- ( -47.30, z-score = -2.39, R) >droGri2.scaffold_15126 1346121 157 - 8399593 -UUUGAUGUGUUAAUUCACUCUGGCUUCUCUUCAGUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAGCACUCUAAUGAGUCUAAACUCAAUUUUUAAA--GCGCCU-GGCGGCAUUUGCAGCCGGGCAAAUUAUGAAUAACAUU -......((((((.((((.....(((((((((((.....((...((((((((.(.......).)))))))).))))))))))))).......(((((....))))).........--..((((-(((.((....)).)))))))......)))))))))). ( -47.10, z-score = -3.29, R) >droMoj3.scaffold_6500 15268547 157 + 32352404 -UUUUCACAGUUUCUUUACUCUGGCUUCUCUUCAGUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAGCACUCUAAUGAGUCUAAAUUGAAUUUUUUGAUUGUGUCU-GACGGCGUUUGCUGUCAGGCCA-UAAAGAAUACAUC- -......((((((..........(((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....))))..)))))).((((((((..((.((((-((((((....))))))))))))-)))))))).....- ( -45.20, z-score = -3.10, R) >droVir3.scaffold_12963 18707870 152 - 20206255 -UUGAAUUGGGCUAUUCACUCUGGCUUCUCUUCAGUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAGCACUCUAAUGAGUCUAAAUUGAAUUUUUUCGU-GUGCCU-GGUGACAUUUGUUGCCGGGCCAAUCAAAAAU------ -(..(.(((((((..........(((((((((((.....((...((((((((.(.......).)))))))).))))))))))))).........))))))).)..)((((((...(-(.((((-((..((....))..))))))))...))))))------ ( -46.61, z-score = -3.26, R) >droWil1.scaffold_180708 11931724 151 + 12563649 -GUGGUGAUGGAAAAUCACUCUGGCUUCUCUUCAGUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGGGCACUCUAAUGAGUCUAAAUUGAAUUUUU-CAUAGUGCCU-GGCGAAA------GUCGGGCCAAUAAUUAAUAAAUU- -......(((((((((((.....(((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....))))......)).))))))-))).(.((((-(((....------))))))))...............- ( -43.40, z-score = -2.58, R) >droPer1.super_10 269404 158 - 3432795 -UUGGCGGUGUAAAGUCACUCUGGCUUCUCUUCAAUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGGGCACUCUAAUGAGUCUAAAAUGAAUUUUUUCGUAGUGCCC-GGCGGCUUCGGUUGCUGGGCCAAUAUUGUAUAAAUU- -...(((((((............(((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....)))).....(((((....))))).(.((((-((((((....))))))))))).))))))).......- ( -49.40, z-score = -2.89, R) >dp4.chr4_group4 2464127 158 - 6586962 -UUGGCGGUGUAAAGUCACUCUGGCUUCUCUUCAAUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGGGCACUCUAAUGAGUCUAAAAUGAAUUUUUUCGUAGUGCCC-GGCGGCUUCGGUUGCUGGGCCAGUAAUGUAUAAAUU- -.((((........))))..((((((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....)))).....(((((....)))))...((((-((((((....))))))))))))).............- ( -48.90, z-score = -2.54, R) >droAna3.scaffold_12916 6573782 158 - 16180835 -AAUACCUGAGGAAAGUACUCUGGCUUCUCUUCAGUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAGCACUCUAAUGAGUCUAAACUAAUUUUUUGUUC-GUGCCCUAGAGGAGUAUUCCUCCGGGGCCAUAAUAAAUAAAUU- -.(((.(((((((((((......)))).))))))).)))((((((((......(((((((.(((..(....)..)))))))).))((((....))))............)))))))-).((((..((((......))))..))))...............- ( -43.80, z-score = -1.96, R) >droEre2.scaffold_4929 3096163 158 + 26641161 CUGGACAUGAA-AU-UUACUCUGGUUUCUCUUCAAUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUAAACAGAAUUUUUGUUUAGAGCCCCGCUGGCGUUUGCCUAUGGGGCCAUAAUUUAUAAAUU- (((((((.(((-((-....(((((((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....)))).....)))))))))))))))).((((((..(((....)))..))))))...............- ( -47.40, z-score = -3.77, R) >droYak2.chr2L 3044797 159 + 22324452 CUGGGUUGGAG-AAGUUACUCUGGUUUCUCUUCAAUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUAAACUGAAUUUUUGCUUAGAGCCCGACUGGCGUAUGCCUCUGGGGCCAUAAAUGAUAAAUU- ((((((..(((-.((((......(((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....))))...))))...)))..)))))).((((.(..(((....)))..).))))...............- ( -41.60, z-score = -0.92, R) >droSec1.super_5 1205977 159 + 5866729 CGGGAGCUGAA-AAGUUACUCUGGUUUCUCUUCAAUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUAAACACAAUUUUUGCUUAGAGCCCCGAUGGCGUUUGCCUUUGGGGCCAUAAAUGAUACAUU- .(((((((...-.)))).)))..(((((((((((.....((...((((((((.(.......).)))))))).))))))))))))).(((....)))(((((.((.......)).)))))(((((((.(((....))).)))))))...............- ( -45.70, z-score = -2.29, R) >droSim1.chr2L 2990141 158 + 22036055 CAGGAGCUGAA-A-GUUACUCUGGUUUCUCUUCAAUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUAAACACAAUUUUUGCUUAGAGCCCCGAUGGCGUUUGCCUUUGGGGCCAUAAAUAAUACAUU- ...((((.(((-(-(((......(((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....))))........))))))))))).(.(((((((.(((....))).))))))))..............- ( -45.30, z-score = -2.66, R) >consensus _UUGGCGUGAA_AAGUUACUCUGGCUUCUCUUCAAUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAGCACUCUAAUGAGUCUAAACUGAAUUUUUGCUUAGUGCCC_GGUGGCGUUUGCCGCUGGGCCAAUAAAUAAUAAAUU_ .......................(((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....)))).......................(((.((.(((....))).))))).................. (-28.17 = -27.07 + -1.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:12:54 2011