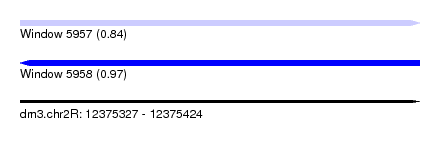
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,375,327 – 12,375,424 |
| Length | 97 |
| Max. P | 0.971548 |

| Location | 12,375,327 – 12,375,424 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 61.58 |
| Shannon entropy | 0.65740 |
| G+C content | 0.36982 |
| Mean single sequence MFE | -21.04 |
| Consensus MFE | -9.68 |
| Energy contribution | -9.84 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.838169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
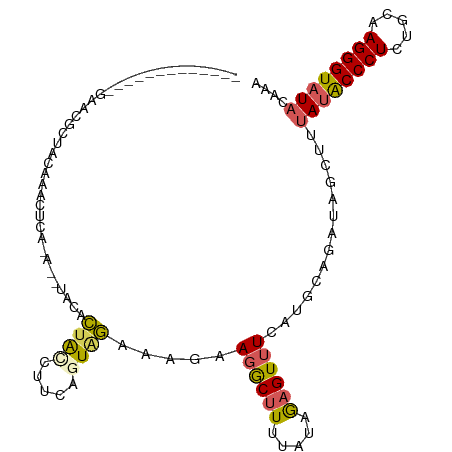
>dm3.chr2R 12375327 97 + 21146708 -------------GAAUGCUAGAAUGUCA-AUUUAGACUAUCUUCUUUGGAAAGAAGACUUUUAUAGAGUUUCAUGCGUAAAGCUUUAUACCCUCUACAAGGGUAUACAAA -------------(((..(((....(((.-.....)))..(((((((....)))))))......)))...)))..((.....))..((((((((.....)))))))).... ( -21.50, z-score = -1.65, R) >droSim1.chr2R 11113978 97 + 19596830 -------------GAAUGCUAGAAUGUCA-AUAUACACUAUCUUCAGUAGAAAGAAGGCUUUCUCAGAGUUUCAAGCAGAUAGCUUUAUACCCUCUGCAAGGGUAUAAUAA -------------...((((.(((..((.-......(((......)))((((((....))))))..))..))).)))).......(((((((((.....)))))))))... ( -22.10, z-score = -1.20, R) >droSec1.super_1 9879248 76 + 14215200 -----------------------------------AGUAGAAGUCAGUAAAAAGAAGGCUUUUUCAGAGUUUCAUGCAUAAAGCUUAAUACCCUCUGCAAGGGUAUAAAAA -----------------------------------.(.(((((((...........))))))).).((((((........)))))).(((((((.....)))))))..... ( -15.50, z-score = -1.37, R) >droYak2.chr2R 4532502 111 - 21139217 AGAGGUGCGAAUCGAACGUUUCCGACUUACACAUACACUACCUUCAGUAGGAAUAACGCAUUCAUGAAGUUUCUUGCAGAUAGCCUUAUACCCUUUGCAAGGGUAUACAAU .(((((((((.(((........)))................(((((....((((.....)))).)))))....)))).....)))))(((((((.....)))))))..... ( -22.20, z-score = -0.62, R) >droEre2.scaffold_4845 9107413 107 - 22589142 AACUGUGCGAAUCGGACGUUUCCGACUUA----UACACUACCUUCAGUAGGAGAAAGGCUUUUAUUAAGUUUCAUGCUGAUAACCUUAAGCCCUUUGCAAGGGUAUACAAU ...((((..(.(((((....))))).)..----))))......((((((.(((....((((.....))))))).)))))).........(((((.....)))))....... ( -23.90, z-score = -0.82, R) >consensus _____________GAACGCUACAAACUCA_A__UACACUACCUUCAGUAGAAAGAAGGCUUUUAUAGAGUUUCAUGCAGAUAGCUUUAUACCCUCUGCAAGGGUAUACAAA .....................................((((.....)))).....((((((.....))))))..............((((((((.....)))))))).... ( -9.68 = -9.84 + 0.16)

| Location | 12,375,327 – 12,375,424 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 61.58 |
| Shannon entropy | 0.65740 |
| G+C content | 0.36982 |
| Mean single sequence MFE | -22.30 |
| Consensus MFE | -10.94 |
| Energy contribution | -11.90 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
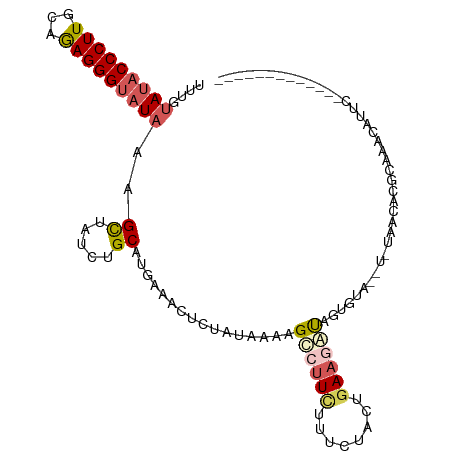
>dm3.chr2R 12375327 97 - 21146708 UUUGUAUACCCUUGUAGAGGGUAUAAAGCUUUACGCAUGAAACUCUAUAAAAGUCUUCUUUCCAAAGAAGAUAGUCUAAAU-UGACAUUCUAGCAUUC------------- ....(((((((((...))))))))).........((..(((...........((((((((....)))))))).(((.....-.))).)))..))....------------- ( -24.00, z-score = -3.04, R) >droSim1.chr2R 11113978 97 - 19596830 UUAUUAUACCCUUGCAGAGGGUAUAAAGCUAUCUGCUUGAAACUCUGAGAAAGCCUUCUUUCUACUGAAGAUAGUGUAUAU-UGACAUUCUAGCAUUC------------- ...((((((((((...)))))))))).(((((((.........((..((((((....))))))...)))))))))......-................------------- ( -21.90, z-score = -1.14, R) >droSec1.super_1 9879248 76 - 14215200 UUUUUAUACCCUUGCAGAGGGUAUUAAGCUUUAUGCAUGAAACUCUGAAAAAGCCUUCUUUUUACUGACUUCUACU----------------------------------- .....((((((((...))))))))...((.....))..(((..((..((((((....))))))...)).)))....----------------------------------- ( -12.50, z-score = -0.89, R) >droYak2.chr2R 4532502 111 + 21139217 AUUGUAUACCCUUGCAAAGGGUAUAAGGCUAUCUGCAAGAAACUUCAUGAAUGCGUUAUUCCUACUGAAGGUAGUGUAUGUGUAAGUCGGAAACGUUCGAUUCGCACCUCU ...((((((.((((((...(((.....)))...))))))...(((((.(((((...)))))....)))))...))))))(((..(((((((....)))))))..))).... ( -28.80, z-score = -1.52, R) >droEre2.scaffold_4845 9107413 107 + 22589142 AUUGUAUACCCUUGCAAAGGGCUUAAGGUUAUCAGCAUGAAACUUAAUAAAAGCCUUUCUCCUACUGAAGGUAGUGUA----UAAGUCGGAAACGUCCGAUUCGCACAGUU .((((((((((((.((((((((((.(((((.((.....))))))).....)))))))).......))))))..)))))----)))((((((....)))))).......... ( -24.31, z-score = -0.85, R) >consensus UUUGUAUACCCUUGCAGAGGGUAUAAAGCUAUCUGCAUGAAACUCUAUAAAAGCCUUCUUUCUACUGAAGAUAGUGUA__U_UAACACGCAAACAUUC_____________ ....(((((((((...)))))))))..((.....))................((((((........))))))....................................... (-10.94 = -11.90 + 0.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:27:25 2011