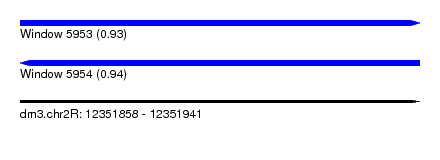
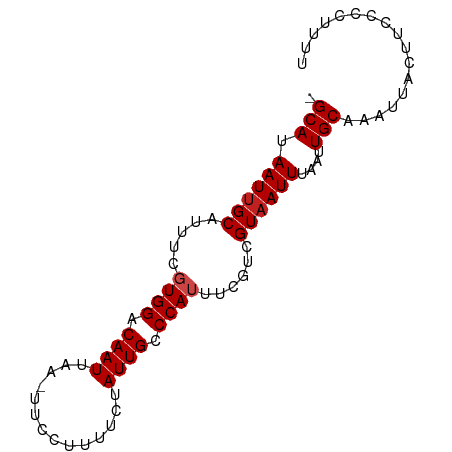
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,351,858 – 12,351,941 |
| Length | 83 |
| Max. P | 0.936518 |

| Location | 12,351,858 – 12,351,941 |
|---|---|
| Length | 83 |
| Sequences | 12 |
| Columns | 84 |
| Reading direction | forward |
| Mean pairwise identity | 94.85 |
| Shannon entropy | 0.10494 |
| G+C content | 0.31986 |
| Mean single sequence MFE | -12.99 |
| Consensus MFE | -12.83 |
| Energy contribution | -12.92 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
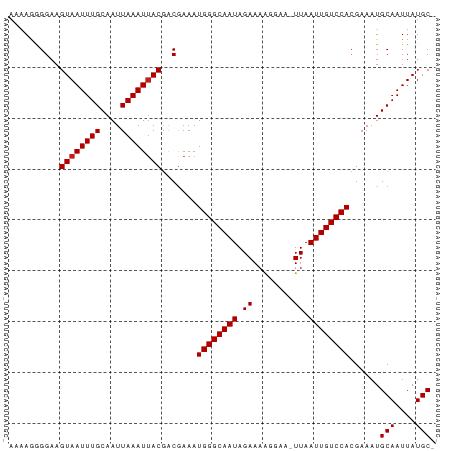
>dm3.chr2R 12351858 83 + 21146708 AAAAGGGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAAGGAA-UUAAUUGUCCACGAAAUGCAAUUAUGCU ..........((((((((....)))))))).......((((((((.((.......-)).))))))))......(((....))). ( -14.00, z-score = -1.56, R) >droSim1.chr2R 11088062 82 + 19596830 AAAAGGGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAAGGAA-UUAAUUGUCCACGAAAUGCAAUUAUGC- ..........((((((((....)))))))).......((((((((.((.......-)).))))))))......(((....)))- ( -12.80, z-score = -1.43, R) >droSec1.super_1 9849319 82 + 14215200 AAAAGGGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAAGGAA-UUAAUUGUCCACGAAAUGCAAUUAUGC- ..........((((((((....)))))))).......((((((((.((.......-)).))))))))......(((....)))- ( -12.80, z-score = -1.43, R) >droYak2.chr2R 4505516 82 - 21139217 AAAAGGGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAAGGAA-UUAAUUGUCCACGAAAUGCAAUUAUGC- ..........((((((((....)))))))).......((((((((.((.......-)).))))))))......(((....)))- ( -12.80, z-score = -1.43, R) >droEre2.scaffold_4845 9082460 82 - 22589142 AAAAGGGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAGGGAA-UUAAUUGUCCACGAAAUGCAAUUAUGC- ..........((((((((....)))))))).......((((((((.((.......-)).))))))))......(((....)))- ( -12.80, z-score = -1.33, R) >droAna3.scaffold_13266 15727799 82 + 19884421 AAAAGGGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAGGGAA-UUAAUUGUCCACGAAAUGCAAUUAUGC- ..........((((((((....)))))))).......((((((((.((.......-)).))))))))......(((....)))- ( -12.80, z-score = -1.33, R) >dp4.chr3 15988299 83 - 19779522 AAGAGGGGAAGUAAUUUGCAAUUAAAUUACAACGAAAUGGGCAAUAGAAAAGGAAAUUAAUUGUCCAAGAAAUGCAAUUAUGC- ..........((((((((....)))))))).......((((((((.((........)).))))))))......(((....)))- ( -13.40, z-score = -2.02, R) >droPer1.super_2 5060982 83 - 9036312 AAGAGGGGAAGUAAUUUGCAAUUAAAUAACAACGAAAUGGGCAAUAGAAAAGGAAAUUAAUUGUCCAAGAAAUGCAAUUAUGC- ..........((((((.((..................((((((((.((........)).))))))))(....)))))))))..- ( -11.30, z-score = -1.29, R) >droWil1.scaffold_180700 5852777 83 - 6630534 ACAAAAGGAAGUAAUUUGCAAUUAAAUUACGGCAAAAUGGGCAAUAGAAAAGAAA-UUAAUUGUCCACGAAAUGCAAUUAUGCU ..........((((((((....)))))))).(((...((((((((.((.......-)).)))))))).....)))......... ( -14.80, z-score = -1.71, R) >droVir3.scaffold_12875 18874824 82 + 20611582 ACAAAAGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAAGAAA-UUAAUUGUCCACGAAAUGCAAUUAUGC- ..........((((((((....)))))))).......((((((((.((.......-)).))))))))......(((....)))- ( -12.80, z-score = -1.57, R) >droMoj3.scaffold_6496 12548082 82 + 26866924 ACAAAAGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAAGAAA-UUAAUUGUCCACGAAAUGCAAUUAUGC- ..........((((((((....)))))))).......((((((((.((.......-)).))))))))......(((....)))- ( -12.80, z-score = -1.57, R) >droGri2.scaffold_15245 4950630 82 + 18325388 ACAAAAGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAAGAAA-UUAAUUGUCCACGAAAUGCAAUUAUGC- ..........((((((((....)))))))).......((((((((.((.......-)).))))))))......(((....)))- ( -12.80, z-score = -1.57, R) >consensus AAAAGGGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAAGGAA_UUAAUUGUCCACGAAAUGCAAUUAUGC_ ..........((((((((....)))))))).......((((((((.((........)).))))))))......(((....))). (-12.83 = -12.92 + 0.08)

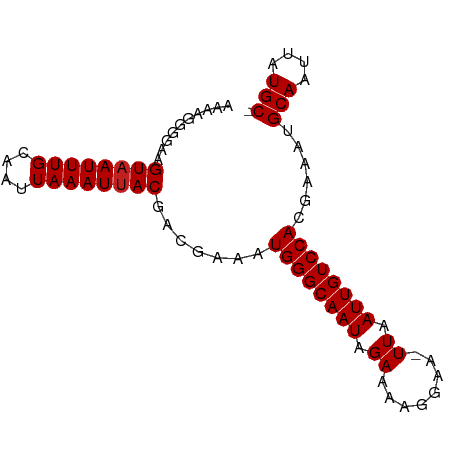
| Location | 12,351,858 – 12,351,941 |
|---|---|
| Length | 83 |
| Sequences | 12 |
| Columns | 84 |
| Reading direction | reverse |
| Mean pairwise identity | 94.85 |
| Shannon entropy | 0.10494 |
| G+C content | 0.31986 |
| Mean single sequence MFE | -11.06 |
| Consensus MFE | -10.30 |
| Energy contribution | -10.55 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.936518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12351858 83 - 21146708 AGCAUAAUUGCAUUUCGUGGACAAUUAA-UUCCUUUUCUAUUGCCCAUUUCGUCGUAAUUUAAUUGCAAAUUACUUCCCCUUUU .(((.((((((((...((((.((((...-..........)))).))))...)).))))))....)))................. ( -11.32, z-score = -2.04, R) >droSim1.chr2R 11088062 82 - 19596830 -GCAUAAUUGCAUUUCGUGGACAAUUAA-UUCCUUUUCUAUUGCCCAUUUCGUCGUAAUUUAAUUGCAAAUUACUUCCCCUUUU -(((.((((((((...((((.((((...-..........)))).))))...)).))))))....)))................. ( -11.12, z-score = -2.21, R) >droSec1.super_1 9849319 82 - 14215200 -GCAUAAUUGCAUUUCGUGGACAAUUAA-UUCCUUUUCUAUUGCCCAUUUCGUCGUAAUUUAAUUGCAAAUUACUUCCCCUUUU -(((.((((((((...((((.((((...-..........)))).))))...)).))))))....)))................. ( -11.12, z-score = -2.21, R) >droYak2.chr2R 4505516 82 + 21139217 -GCAUAAUUGCAUUUCGUGGACAAUUAA-UUCCUUUUCUAUUGCCCAUUUCGUCGUAAUUUAAUUGCAAAUUACUUCCCCUUUU -(((.((((((((...((((.((((...-..........)))).))))...)).))))))....)))................. ( -11.12, z-score = -2.21, R) >droEre2.scaffold_4845 9082460 82 + 22589142 -GCAUAAUUGCAUUUCGUGGACAAUUAA-UUCCCUUUCUAUUGCCCAUUUCGUCGUAAUUUAAUUGCAAAUUACUUCCCCUUUU -(((.((((((((...((((.((((...-..........)))).))))...)).))))))....)))................. ( -11.12, z-score = -2.28, R) >droAna3.scaffold_13266 15727799 82 - 19884421 -GCAUAAUUGCAUUUCGUGGACAAUUAA-UUCCCUUUCUAUUGCCCAUUUCGUCGUAAUUUAAUUGCAAAUUACUUCCCCUUUU -(((.((((((((...((((.((((...-..........)))).))))...)).))))))....)))................. ( -11.12, z-score = -2.28, R) >dp4.chr3 15988299 83 + 19779522 -GCAUAAUUGCAUUUCUUGGACAAUUAAUUUCCUUUUCUAUUGCCCAUUUCGUUGUAAUUUAAUUGCAAAUUACUUCCCCUCUU -(((.(((((((.....(((.((((..(........)..)))).)))......)))))))....)))................. ( -12.40, z-score = -3.07, R) >droPer1.super_2 5060982 83 + 9036312 -GCAUAAUUGCAUUUCUUGGACAAUUAAUUUCCUUUUCUAUUGCCCAUUUCGUUGUUAUUUAAUUGCAAAUUACUUCCCCUCUU -(((.(((.(((.....(((.((((..(........)..)))).)))......))).)))....)))................. ( -8.80, z-score = -1.44, R) >droWil1.scaffold_180700 5852777 83 + 6630534 AGCAUAAUUGCAUUUCGUGGACAAUUAA-UUUCUUUUCUAUUGCCCAUUUUGCCGUAAUUUAAUUGCAAAUUACUUCCUUUUGU .(((.((((((.....((((.((((...-..........)))).))))......))))))....)))................. ( -11.22, z-score = -0.90, R) >droVir3.scaffold_12875 18874824 82 - 20611582 -GCAUAAUUGCAUUUCGUGGACAAUUAA-UUUCUUUUCUAUUGCCCAUUUCGUCGUAAUUUAAUUGCAAAUUACUUCCUUUUGU -(((.((((((((...((((.((((...-..........)))).))))...)).))))))....)))................. ( -11.12, z-score = -1.55, R) >droMoj3.scaffold_6496 12548082 82 - 26866924 -GCAUAAUUGCAUUUCGUGGACAAUUAA-UUUCUUUUCUAUUGCCCAUUUCGUCGUAAUUUAAUUGCAAAUUACUUCCUUUUGU -(((.((((((((...((((.((((...-..........)))).))))...)).))))))....)))................. ( -11.12, z-score = -1.55, R) >droGri2.scaffold_15245 4950630 82 - 18325388 -GCAUAAUUGCAUUUCGUGGACAAUUAA-UUUCUUUUCUAUUGCCCAUUUCGUCGUAAUUUAAUUGCAAAUUACUUCCUUUUGU -(((.((((((((...((((.((((...-..........)))).))))...)).))))))....)))................. ( -11.12, z-score = -1.55, R) >consensus _GCAUAAUUGCAUUUCGUGGACAAUUAA_UUCCUUUUCUAUUGCCCAUUUCGUCGUAAUUUAAUUGCAAAUUACUUCCCCUUUU .(((.((((((.....((((.((((..............)))).))))......))))))....)))................. (-10.30 = -10.55 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:27:22 2011