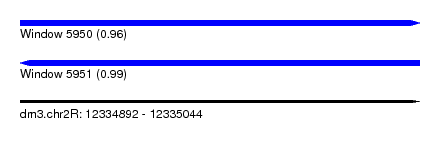
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,334,892 – 12,335,044 |
| Length | 152 |
| Max. P | 0.992845 |

| Location | 12,334,892 – 12,335,044 |
|---|---|
| Length | 152 |
| Sequences | 6 |
| Columns | 184 |
| Reading direction | forward |
| Mean pairwise identity | 79.41 |
| Shannon entropy | 0.35289 |
| G+C content | 0.42584 |
| Mean single sequence MFE | -46.87 |
| Consensus MFE | -30.07 |
| Energy contribution | -30.25 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
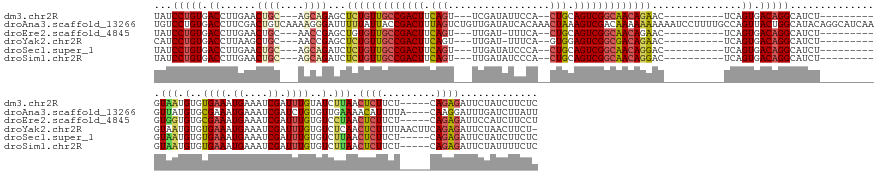
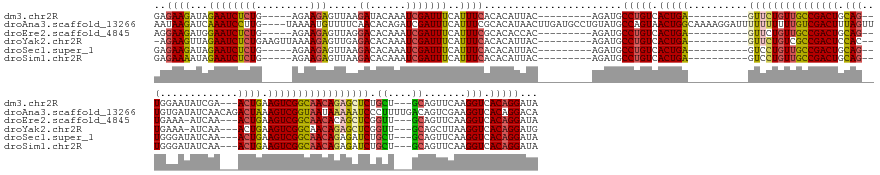
>dm3.chr2R 12334892 152 + 21146708 UAUCCUGUGACCUUGAACUGC---AGCAGAGCUCUGUUGCCGACUUCAGU---UCGAUAUUCCA--CUGCAGUCGGCAACAGAAC----------UCAGUGACAGGCAUCU---------GUAAUGUGUGAAAUGAAAUCGAUUUGUAUCUUAACUCUUCU-----CAGAGAUUCUAUCUUCUC ...(((((.((.(((.....)---))..(((.(((((((((((((.((((---..(.....).)--))).))))))))))))).)----------)).)).))))).....---------.(((.(((..((.((....)).))..))).))).((((...-----.))))............. ( -45.40, z-score = -2.18, R) >droAna3.scaffold_13266 15710357 180 + 19884421 UGUCCUGUGACCUUCGACUGUCAAAAGGGAUUUUUAUUACCGACUUUAGUCUGUUGAUAUCACAAACUAAAGUCGACAAAAAAAAAAUCCUUUUGCCAGUUACUGGCAUACAGGCAUCAAGUUAUGUGCGAAAUGAAAUCGAUCUGUGUUGAAAACAUUUUA----CAAGGAUUUGAUCUUAUU .(.((((((.((...(((((.((((((((.(((((.((..((((((((((.(((.(....)))).))))))))))...)).))))).)))))))).)))))...))..)))))))((((((((...((..(((((...(((((....)))))...)))))..----))..))))))))...... ( -55.80, z-score = -5.31, R) >droEre2.scaffold_4845 9065451 151 - 22589142 UAUCCUGUGACCUUGAACUGC---AACCGAGCUGUGUUGCCGACUUCAGU---UUGAU-UUUCA--CUGCAGUCGGCAACAGAAC----------UCAGUGACAGGCAUCU---------GUGGUGUGCGAAAUGAAAUCGAUUUGUGUCCUAACUCUUCU-----CAGAGAUUCCAUCUUCCU ...(((((.((.(((.....)---))..(((.(.(((((((((((.((((---.....-....)--))).))))))))))).).)----------)).)).))))).....---------.(((..((((((.((....)).))))))..))).((((...-----.))))............. ( -44.70, z-score = -1.56, R) >droYak2.chr2R 4488101 155 - 21139217 CAUCCUGUGACCUUAAGCUGC---AACCGAGCUCUGUUGCCGACUUCAGU---UUGAU-UUUCA--GUGGAGUCGGCGACAGAAC----------UCAGUGACAGGCAUCU---------GUAAUGUGUGAAAUGAAAUCGAUUUGUGUCUCAACUCUUUUAACUUCAGAGAUUCUAACUUCU- ....((.(((..(((((((..---.((.(((.((((((((((((((((..---(((..-...))--))))))))))))))))).)----------)).))....)))....---------......((.((.(..((.....))..).)).))......))))..))).))............- ( -45.20, z-score = -1.53, R) >droSec1.super_1 9832474 152 + 14215200 UAUCCUGUGACCUUGAACUGC---AGCAGAUCUCUGUUGCCGACUUCAGU---UUGAUAUCCCA--CUGCAGUCGGCAACAGGAC----------UCAGUGACAGGCAUCU---------GUAAUGUGUGAAAUGAAAUCGAUUUGUGUCUUAACUCUUCU-----CAGAGAUUCUAUCUUCUC .((((((.((.........((---(.(((((((((((((((((((.((((---..(....)..)--))).)))))))))))))((----------.((...((((....))---------))..)).))...........))))))))).........)).-----))).)))........... ( -45.07, z-score = -1.79, R) >droSim1.chr2R 11070835 152 + 19596830 UAUCCUGUGACCUUGAACUGC---AGCAGAUCUCUGUUGCCGACUUCAGU---UUGAUAUCCCA--CUGCAGUCGGCAACAGGAC----------UCAGUGACAGGCAUCU---------GUAAUGUGUGAAAUGAAAUCGAUUUGUGUCUUAACUCUUCU-----CAGAGAUUCUAUUUUCUC .((((((.((.........((---(.(((((((((((((((((((.((((---..(....)..)--))).)))))))))))))((----------.((...((((....))---------))..)).))...........))))))))).........)).-----))).)))........... ( -45.07, z-score = -1.91, R) >consensus UAUCCUGUGACCUUGAACUGC___AACAGAGCUCUGUUGCCGACUUCAGU___UUGAUAUCCCA__CUGCAGUCGGCAACAGAAC__________UCAGUGACAGGCAUCU_________GUAAUGUGUGAAAUGAAAUCGAUUUGUGUCUUAACUCUUCU_____CAGAGAUUCUAUCUUCUC ...(((((.((......(((......)))...(((((((((((((.(((.................))).)))))))))))))...............)).))))).......................((((((.(((((((....)))........(((......))))))).))).))).. (-30.07 = -30.25 + 0.17)
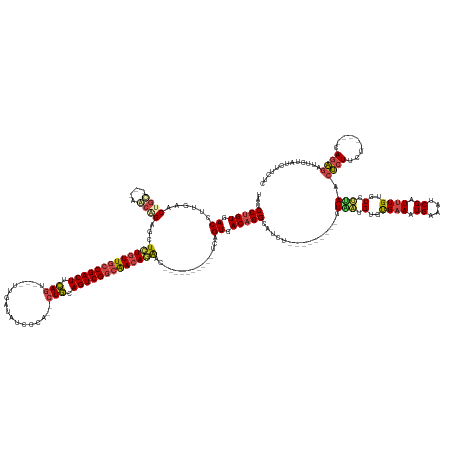
| Location | 12,334,892 – 12,335,044 |
|---|---|
| Length | 152 |
| Sequences | 6 |
| Columns | 184 |
| Reading direction | reverse |
| Mean pairwise identity | 79.41 |
| Shannon entropy | 0.35289 |
| G+C content | 0.42584 |
| Mean single sequence MFE | -50.15 |
| Consensus MFE | -31.94 |
| Energy contribution | -32.42 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.992845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
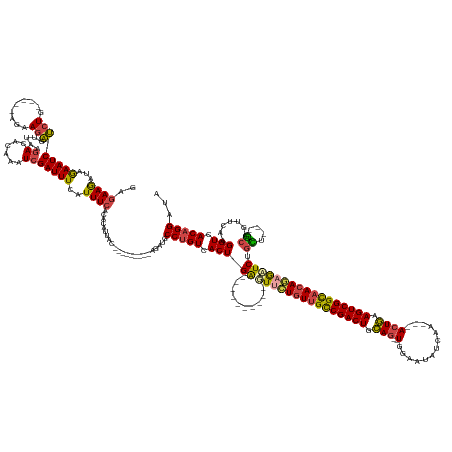
>dm3.chr2R 12334892 152 - 21146708 GAGAAGAUAGAAUCUCUG-----AGAAGAGUUAAGAUACAAAUCGAUUUCAUUUCACACAUUAC---------AGAUGCCUGUCACUGA----------GUUCUGUUGCCGACUGCAG--UGGAAUAUCGA---ACUGAAGUCGGCAACAGAGCUCUGCU---GCAGUUCAAGGUCACAGGAUA ((((........))))((-----(((.(((....(((....)))...))).)))))........---------.....(((((.(((((----------(((((((((((((((.(((--(..........---)))).))))))))))))))))).((.---...))....))).)))))... ( -52.50, z-score = -3.23, R) >droAna3.scaffold_13266 15710357 180 - 19884421 AAUAAGAUCAAAUCCUUG----UAAAAUGUUUUCAACACAGAUCGAUUUCAUUUCGCACAUAACUUGAUGCCUGUAUGCCAGUAACUGGCAAAAGGAUUUUUUUUUUGUCGACUUUAGUUUGUGAUAUCAACAGACUAAAGUCGGUAAUAAAAAUCCCUUUUGACAGUCGAAGGUCACAGGACA ......(((((.....((----..(((((...((..........))...)))))..))......))))).(((((..(((..(.((((.(((((((((((((...((..((((((((((((((.......))))))))))))))..)).)))))).))))))).)))).)..))).)))))... ( -56.00, z-score = -5.47, R) >droEre2.scaffold_4845 9065451 151 + 22589142 AGGAAGAUGGAAUCUCUG-----AGAAGAGUUAGGACACAAAUCGAUUUCAUUUCGCACACCAC---------AGAUGCCUGUCACUGA----------GUUCUGUUGCCGACUGCAG--UGAAA-AUCAA---ACUGAAGUCGGCAACACAGCUCGGUU---GCAGUUCAAGGUCACAGGAUA .(..(((((((((((((.-----...)))((....)).......))))))))))..)..(((..---------.((...((((.(((((----------(((.(((((((((((.(((--(....-.....---)))).))))))))))).)))))))).---)))).))..)))......... ( -52.30, z-score = -2.85, R) >droYak2.chr2R 4488101 155 + 21139217 -AGAAGUUAGAAUCUCUGAAGUUAAAAGAGUUGAGACACAAAUCGAUUUCAUUUCACACAUUAC---------AGAUGCCUGUCACUGA----------GUUCUGUCGCCGACUCCAC--UGAAA-AUCAA---ACUGAAGUCGGCAACAGAGCUCGGUU---GCAGCUUAAGGUCACAGGAUG -..........(((.(((..((.(((.(((((((........)))))))..))).)).......---------....((.(((.(((((----------(((((((.(((((((.((.--((...-..)).---..)).))))))).)))))))))))).---))))).........)))))). ( -47.30, z-score = -2.29, R) >droSec1.super_1 9832474 152 - 14215200 GAGAAGAUAGAAUCUCUG-----AGAAGAGUUAAGACACAAAUCGAUUUCAUUUCACACAUUAC---------AGAUGCCUGUCACUGA----------GUCCUGUUGCCGACUGCAG--UGGGAUAUCAA---ACUGAAGUCGGCAACAGAGAUCUGCU---GCAGUUCAAGGUCACAGGAUA ((((........))))((-----(((.(((....((......))...))).)))))........---------.....(((((.(((..----------...((((((((((((.(((--(..(....)..---)))).)))))))))))).((.(((..---.))).))..))).)))))... ( -46.40, z-score = -1.63, R) >droSim1.chr2R 11070835 152 - 19596830 GAGAAAAUAGAAUCUCUG-----AGAAGAGUUAAGACACAAAUCGAUUUCAUUUCACACAUUAC---------AGAUGCCUGUCACUGA----------GUCCUGUUGCCGACUGCAG--UGGGAUAUCAA---ACUGAAGUCGGCAACAGAGAUCUGCU---GCAGUUCAAGGUCACAGGAUA ((((........))))((-----(((.(((....((......))...))).)))))........---------.....(((((.(((..----------...((((((((((((.(((--(..(....)..---)))).)))))))))))).((.(((..---.))).))..))).)))))... ( -46.40, z-score = -1.93, R) >consensus GAGAAGAUAGAAUCUCUG_____AGAAGAGUUAAGACACAAAUCGAUUUCAUUUCACACAUUAC_________AGAUGCCUGUCACUGA__________GUUCUGUUGCCGACUGCAG__UGGAAUAUCAA___ACUGAAGUCGGCAACAGAGAUCUGCU___GCAGUUCAAGGUCACAGGAUA ...........(((.(((..........................((.......))......................((((...((((...........(((((((((((((((.(((..(((....))).....))).)))))))))))))))..........))))...))))..)))))). (-31.94 = -32.42 + 0.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:27:20 2011