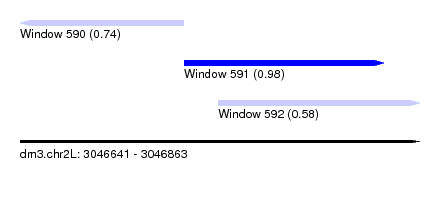
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,046,641 – 3,046,863 |
| Length | 222 |
| Max. P | 0.980119 |

| Location | 3,046,641 – 3,046,732 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 74.54 |
| Shannon entropy | 0.41923 |
| G+C content | 0.49850 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -12.23 |
| Energy contribution | -14.60 |
| Covariance contribution | 2.37 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.739967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
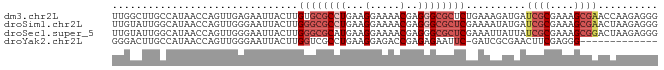
>dm3.chr2L 3046641 91 - 23011544 UUGGCUUGCCAUAACCAGUUGAGAAUUACUUGUGCGCCUGAAGGAAAACGAGGGCGCUCUGAAAGAUGAUCGCGAAAGCGAACCAAGAGGG .(((....)))...((..(((.......(((..((((((...(.....)..)))))).....)))....((((....))))..)))..)). ( -27.10, z-score = -1.70, R) >droSim1.chr2L 2987681 91 - 22036055 UUGUAUUGGCAUAACCAGUUGGGAAUUACUUGGGCGCCUGAAGGAAAACGAGGGCGCUCGAAAAUAUGAUCGCGAAAGCGAACUAAGAGGG ..............((..((((.......((((((((((...(.....)..))))))))))........((((....)))).))))..)). ( -30.70, z-score = -3.87, R) >droSec1.super_5 1203926 91 - 5866729 UUGUAUUGGCAUAACCAGUUGGGAAUUACUUGGGCGCAUGAAGGAAAACGAGGGCGCUCGAAAUUAUUAUCGCGAAAGCGGACUAAGAGGG ..............((..((((.......((((((((.....(.....)....))))))))........((((....)))).))))..)). ( -24.50, z-score = -2.22, R) >droYak2.chr2L 3042208 77 - 22324452 GGGACUUGCCAUAACCAGUUGGGAAUUACUUGGUCGCCUGAAGGAGACCGAGAGAAUUC-GAUCGCGAACUUCGAGGG------------- (((..((((.....((....))(((((.(((((((.((....)).)))))))..)))))-....))))..))).....------------- ( -21.90, z-score = -0.50, R) >consensus UUGUAUUGCCAUAACCAGUUGGGAAUUACUUGGGCGCCUGAAGGAAAACGAGGGCGCUCGAAAAGAUGAUCGCGAAAGCGAACUAAGAGGG .............................((((((((((...(.....)..))))))))))........((((....)))).......... (-12.23 = -14.60 + 2.37)
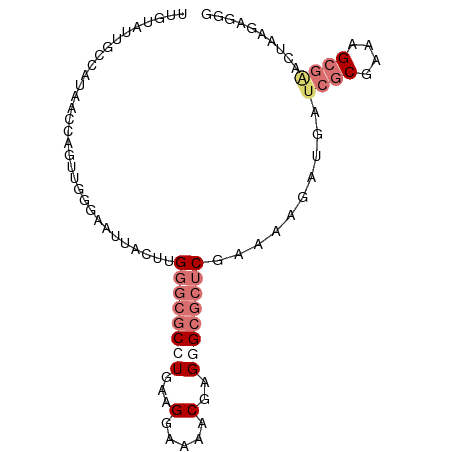

| Location | 3,046,732 – 3,046,843 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.15 |
| Shannon entropy | 0.42645 |
| G+C content | 0.52323 |
| Mean single sequence MFE | -40.96 |
| Consensus MFE | -23.84 |
| Energy contribution | -24.36 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3046732 111 + 23011544 CGUAGA-AUCCCCAUCGAGUGUCGGGGAUCAUCAUGUCU----CGAUCGCCGCUUCAGUU-GUGGAGCGAGAGCUUACGCAAUGGAGCGGAGUGAUGAGCACAUUAUCCGAGGCAAU ....((-.(((((..(....)..)))))))....(((((----((....((((((((.((-(((((((....)))).))))))))))))).((((((....)))))).))))))).. ( -46.80, z-score = -3.42, R) >droSim1.chr2L 2987772 114 + 22036055 CGUAGA-GUCCCCAUCGAGUGUAGGGGAUCAUUUUAUCUAUCUCGAUCGCCGCUUCAGUU-GUGGAGCGAGAGCUUACGCAAUGGAGCGGAGUGAUGAGUA-GUUAUCCGAGGCAAU .(((((-((((((..........)))))).......)))))((((....((((((((.((-(((((((....)))).)))))))))))))...(((((...-.)))))))))..... ( -41.21, z-score = -2.52, R) >droSec1.super_5 1204017 114 + 5866729 CGUAGA-AUCCCCAUCGAGUGCCGGGGAUCAUUCUAUCUAUCUCGAUCGCCGCUUCAGUU-GUGGAGCGAGAGCUUACGCAAUGGAGCGGAGUGAUGAGUA-GUUAUCCGAGGCAAU .(((((-((((((..........))))))...)))))...(((((....((((((((.((-(((((((....)))).)))))))))))))...(((((...-.)))))))))).... ( -42.70, z-score = -2.15, R) >droYak2.chr2L 3042285 111 + 22324452 AGUAGA-ACCCCCAUCGACGACUGUGGAUCAGCAUGUCU----CGAUCGCCGCUUCAGUU-GUGGAGCGAGAGCUUACGCAAUGGAGCGGAGUGAUGAGCACAUUAUCCGAGGCAAU ......-......(((((.((((((......))).))))----)))).(((((((((.((-(((((((....)))).)))))))))))(((.(((((....))))))))..)))... ( -41.10, z-score = -2.27, R) >droAna3.scaffold_12916 6571790 110 - 16180835 UGUAGACAGGCCCCUCGGUUUCAACG--UCAAGUCGACU-CCAAACUCCCCUCGUCAGUUUGUGGAACACCAAUUC-CGCAUUGAUGUUUGGGACUGAGUA---UAUCCGAGGCAAU ............((((((.......(--((.....))).-....(((((((.((((((..(((((((......)))-))))))))))...)))...)))).---...)))))).... ( -33.00, z-score = -1.52, R) >consensus CGUAGA_AUCCCCAUCGAGUGCAGGGGAUCAUCAUGUCU__C_CGAUCGCCGCUUCAGUU_GUGGAGCGAGAGCUUACGCAAUGGAGCGGAGUGAUGAGUA_AUUAUCCGAGGCAAU ...........((.(((...........((((((((((......)))..((((((((....(((((((....)))).)))..)))))))).)))))))..........))))).... (-23.84 = -24.36 + 0.52)

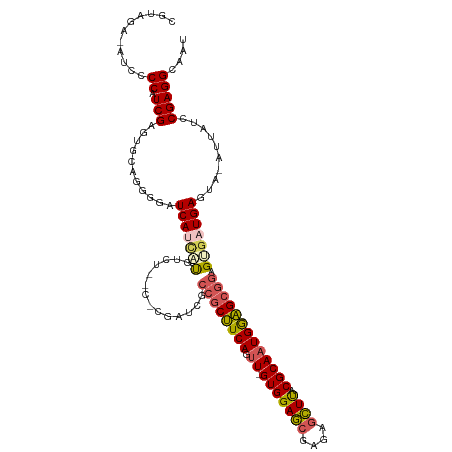
| Location | 3,046,751 – 3,046,863 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.48 |
| Shannon entropy | 0.39431 |
| G+C content | 0.51582 |
| Mean single sequence MFE | -40.70 |
| Consensus MFE | -18.74 |
| Energy contribution | -19.98 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.583070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3046751 112 + 23011544 GUCGGGGAUCA----UCAUGUCUCGAUCGCCGCU--UCAGUU-GUGGAGCGAGAGCUUACGCAAUGGAGCGGAGUGAUGAGCACAUUAUCCGAGGCAAUUUUUUUAGUGCCUGGCCGCG (.(((((((..----..(((((((.(((.(((((--(((.((-(((((((....)))).)))))))))))))...)))))).)))).)))).(((((..........)))))..)))). ( -45.10, z-score = -2.55, R) >droSim1.chr2L 2987791 115 + 22036055 GUAGGGGAUCAUUUUAUCUAUCUCGAUCGCCGCU--UCAGUU-GUGGAGCGAGAGCUUACGCAAUGGAGCGGAGUGAUGAGUA-GUUAUCCGAGGCAAUUUUUUUAGUGCCUGGCCGCG ...(.((..........(((.(((.(((.(((((--(((.((-(((((((....)))).)))))))))))))...))))))))-)....((.(((((..........))))))))).). ( -41.00, z-score = -2.03, R) >droSec1.super_5 1204036 115 + 5866729 GCCGGGGAUCAUUCUAUCUAUCUCGAUCGCCGCU--UCAGUU-GUGGAGCGAGAGCUUACGCAAUGGAGCGGAGUGAUGAGUA-GUUAUCCGAGGCAAUUUUUUUAGUGCCUGGCCGCG (((..((((....(((..((((.(.....(((((--(((.((-(((((((....)))).))))))))))))).).))))..))-)..)))).(((((..........)))))))).... ( -45.00, z-score = -2.37, R) >droYak2.chr2L 3042304 112 + 22324452 ACUGUGGAUCA----GCAUGUCUCGAUCGCCGCU--UCAGUU-GUGGAGCGAGAGCUUACGCAAUGGAGCGGAGUGAUGAGCACAUUAUCCGAGGCAAUUUUUUUAGUGCCUAGCCGCA ...((((....----((((((((((....(((((--(((.((-(((((((....)))).))))))))))))).((((((....)))))).)))))))..........)))....)))). ( -44.10, z-score = -2.74, R) >droAna3.scaffold_12916 6571810 110 - 16180835 --UCAACGUCA---AGUCGACUCCAAACUCCCCUCGUCAGUUUGUGGAACACCAA-UUCCGCAUUGAUGUUUGGGACUGAGUA---UAUCCGAGGCAAUUUUUUGAGUACCCGGCUGCA --.....((..---(((((((((.....((((..((((((..(((((((......-)))))))))))))...))))..)))).---((..(((((.....)))))..))..))))))). ( -28.30, z-score = -0.60, R) >consensus GCCGGGGAUCA____AUCUAUCUCGAUCGCCGCU__UCAGUU_GUGGAGCGAGAGCUUACGCAAUGGAGCGGAGUGAUGAGUA_AUUAUCCGAGGCAAUUUUUUUAGUGCCUGGCCGCG ...(.((..............((((..(.(((((..(((....(((((((....)))).)))..)))))))).)...))))........(((.((((..........))))))))).). (-18.74 = -19.98 + 1.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:12:52 2011