| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,308,357 – 12,308,430 |
| Length | 73 |
| Max. P | 0.530044 |

| Location | 12,308,357 – 12,308,430 |
|---|---|
| Length | 73 |
| Sequences | 12 |
| Columns | 81 |
| Reading direction | forward |
| Mean pairwise identity | 72.10 |
| Shannon entropy | 0.57518 |
| G+C content | 0.51856 |
| Mean single sequence MFE | -20.72 |
| Consensus MFE | -10.65 |
| Energy contribution | -10.75 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.18 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.530044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
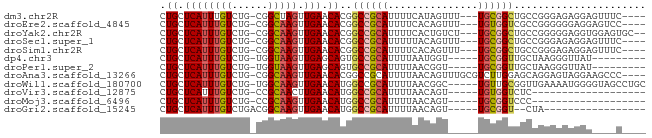
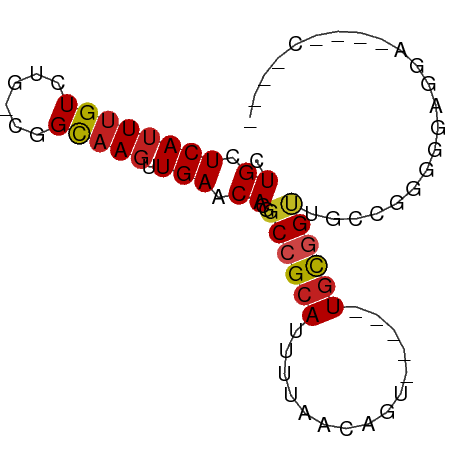
>dm3.chr2R 12308357 73 + 21146708 CUGCUCAUUUGUCUG-CGGCUAGUUGAACACGGCCGCAUUUUCAUAGUUU---UGCGGCUGCCGGGAGAGGAGUUUC---- ..((((...(.(((.-((((..((.....))(((((((............---)))))))))))))).).))))...---- ( -23.00, z-score = -0.80, R) >droEre2.scaffold_4845 9037320 73 - 22589142 CUGCUCAUUUGUCUG-CGGCAAGUUGAACACGGCCGCAUUUUCACAGUUU---UGUGGUCGCCGGGGGGAGGAGUCC---- ..((((...(.(((.-((((..((.....))(((((((............---))))))))))).))).).))))..---- ( -24.30, z-score = -0.73, R) >droYak2.chr2R 4460855 75 - 21139217 CUGCUCAUUUGUCUG-CGGCAAGUUGAACACGGCCGCAUUUUCACUGUCU---UGCGGCUGCCGGGGGAGGUGGAGUGC-- ..((((.....(((.-((((..((.....))(((((((............---))))))))))).))).....))))..-- ( -27.40, z-score = -0.60, R) >droSec1.super_1 9805994 73 + 14215200 CUGCUCAUUUGUCUG-CGGCAAGUUGAACACGGCCGCAUUUUUACAGUUU---UGCGGCUGCCGGGAGAGGAGUUUC---- ..((((...(.(((.-((((..((.....))(((((((............---)))))))))))))).).))))...---- ( -23.50, z-score = -0.98, R) >droSim1.chr2R 11041212 73 + 19596830 CUGCUCAUUUGUCUG-CGGCAAGUUGAACACGGCCGCAUUUUCACAGUUU---UGCGGCUGCCGGGAGAGGAGUUUC---- ..((((...(.(((.-((((..((.....))(((((((............---)))))))))))))).).))))...---- ( -23.50, z-score = -0.73, R) >dp4.chr3 15943067 66 - 19779522 CUGCUCAUUUGUCUG-UGGUAAGUUGAGCAGUGCCGCAUUUUAAUGGU-----UGCGGUUGCUAAGGGUUAU--------- ((((((((((..(..-.)..))).))))))).((((((..........-----)))))).............--------- ( -18.50, z-score = -1.23, R) >droPer1.super_2 5015849 66 - 9036312 CUGCUCAUUUGUCUG-UGGUAAGUUGAGCAGUGCCGCAUUUUAACGGU-----UGCGGUUGCUAAGGGUUAU--------- ((((((((((..(..-.)..))).))))))).((((((..........-----)))))).............--------- ( -18.50, z-score = -0.98, R) >droAna3.scaffold_13266 15683563 76 + 19884421 CUGCUCAUUUGUCUG-CGGCAAGUUGAACACGGCCGCAUUUUAACAGUUUGCGUCUUGGAGCAGGAGUAGGAAGCCC---- ((((((.......((-((((..(.....)...))))))..........((((........)))))))))).......---- ( -21.60, z-score = -0.01, R) >droWil1.scaffold_180700 730240 75 + 6630534 CUGCUCAUUUGUCUG-UGGCAAGUUGAACAUGGCCGCAUUUUAACGGC-----UGUUGCGGUUGAAAAUGGGGUAGCCUGC ((((((.......((-((((..(.....)...))))))(((((((.((-----....)).)))))))...))))))..... ( -23.70, z-score = -1.03, R) >droVir3.scaffold_12875 18822999 56 + 20611582 CUGCUCAUUUGUCUG-CCGCAACUUGAACAUGGCCGCAUUUUAACAGU-----UGUGGUCUC------------------- ..............(-((((((((..((.(((....))).))...)))-----))))))...------------------- ( -12.40, z-score = -0.60, R) >droMoj3.scaffold_6496 12490617 56 + 26866924 CUGCUCAUUUGUCUG-CCGCAAGUUGAACAUGGCCGCAUUUUAACAGU-----UGCGGUCCC------------------- .((.((((((((...-..))))).))).)).(((((((..........-----)))))))..------------------- ( -14.50, z-score = -0.89, R) >droGri2.scaffold_15245 4902163 57 + 18325388 CUGCUCAUUUGUCUGACGGCAAGUUGAACAUGGCCGCAUUUUAACAGU-----UGCGGU--CUA----------------- .((.(((((((((....)))))).))).)).(((((((..........-----))))))--)..----------------- ( -17.70, z-score = -2.02, R) >consensus CUGCUCAUUUGUCUG_CGGCAAGUUGAACACGGCCGCAUUUUAACAGU_____UGCGGUUGCCGGGGGAGGA____C____ .((.(((((((((....)))))).))).))..((((((...............))))))...................... (-10.65 = -10.75 + 0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:27:17 2011