
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,301,902 – 12,301,997 |
| Length | 95 |
| Max. P | 0.682694 |

| Location | 12,301,902 – 12,301,997 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.35 |
| Shannon entropy | 0.51010 |
| G+C content | 0.44158 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -14.70 |
| Energy contribution | -13.79 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.682694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12301902 95 + 21146708 AGCCACGCAGCCAUCGACAGAGAGCAAAUAUUUUGACAGCUGUCAGUCGAGGGAAAUGAUUUCGCAUUUCUGUU-----GCUUCCACUUUUUCC-CGGGGC---------- .(((..(((((..(((((.((.(((.............))).)).))))).(((((((......))))))))))-----))..((.........-.)))))---------- ( -27.92, z-score = -1.35, R) >droSim1.chr2R 11034836 95 + 19596830 AGCCACGCAGCCAUCGUAAGAGAGCAAAUAUUUUGACAGCUGUCAGUCGAGGGAAAUGAUUUCGCAUUUCUGUU-----GCUUCCACUUUUUCC-CGGGGC---------- .(((.(((.((..((....))..))........((((....))))).)).((((((.......(((.......)-----))........)))))-)..)))---------- ( -23.86, z-score = -0.08, R) >droSec1.super_1 9799581 95 + 14215200 AGCCACGCAGCCAUCGACAGAGAGCAAAUAUUUUGACAGCUGUCAGUCGAGGGAAAUGAUUUCGCAUUUCUGUU-----GCUUCCACUUUUUCC-CGGGGC---------- .(((..(((((..(((((.((.(((.............))).)).))))).(((((((......))))))))))-----))..((.........-.)))))---------- ( -27.92, z-score = -1.35, R) >droYak2.chr2R 4454480 96 - 21139217 AGCCACGCAGCCAUCGACAGAGAGCAAAUAUUUUGACAGCUGUCAGUCGAGGGAAAUGAUUUCGCAUUUCUGUU-----GCUUCCACUUUUUCCAUGGGGC---------- .(((..(((((..(((((.((.(((.............))).)).))))).(((((((......))))))))))-----))..(((.........))))))---------- ( -28.72, z-score = -1.59, R) >droEre2.scaffold_4845 9030943 95 - 22589142 AGCCACGCAGCCAUCGACAGAGAGCAAAUAUUUUGACAGCUGUCAGUCGAGGGAAAUGAUUUCGCAUUUCUGUU-----GCUUCCACUUUUUCC-CGGGGC---------- .(((..(((((..(((((.((.(((.............))).)).))))).(((((((......))))))))))-----))..((.........-.)))))---------- ( -27.92, z-score = -1.35, R) >droAna3.scaffold_13266 15677159 95 + 19884421 AGCCACGCAACCAUCGAAGGAGAGCAAAUAUUUUGACAGCUGUCAGUCGAGGGAAAUGAUUUCGCAUUUCUGUU-----GCUUCCCAUUUUUGC-UGGGGC---------- .(((..(((((..((((..((.(((.............))).))..)))).(((((((......))))))))))-----))...(((.......-))))))---------- ( -28.72, z-score = -1.27, R) >dp4.chr3 15936767 102 - 19779522 AGGGACGCA--CUUUGACAGAGAGCAAAUAUUUUGACAGCUGUCAGUUGAAGAAAAUGAUUUCACAUUUUUGCC------UUGCCUUUUUU-AUGCACCAUCUCCUGCCCC .(((..(((--(((..((.((.(((.............))).)).))..)))((((((......))))))))).------...........-..(((........)))))) ( -19.92, z-score = -0.19, R) >droPer1.super_2 5009533 103 - 9036312 AGGGACGCA--CUUUGACAGAGAGCAAAUAUUUUGACAGCUGUCAGUUGAAGAAAAUGAUUUCACAUUUUUGCC------UUGCCUUUUUUUAUGCACCAUCUCCUGCCCC .(((..(((--(((..((.((.(((.............))).)).))..)))((((((......))))))))).------..............(((........)))))) ( -19.92, z-score = -0.18, R) >droVir3.scaffold_12875 18817694 88 + 20611582 AACAGCGCA--CGUUGACAGAGAGCAAAUAUUUUGACAGCUGUCAGUUGAAGAAAAUGAUUUUACAUUUUCCUU------CUUUUUUUUUUCCUCC--------------- .........--..(((((((....(((.....)))....)))))))..((((((((((......)))))).)))------)...............--------------- ( -16.20, z-score = -1.08, R) >droMoj3.scaffold_6496 12485200 94 + 26866924 ---CGCGCGCA-GUUGACAGAGAGCAAAUAUUUUGACAGCUGUCAGUUGAAGAAAAUGAUUUUACAUAUUUUUUUCGA-GCUGCCGCUGCCUUCUUUGU------------ ---.(((.(((-((((((((....(((.....)))....)))))).((((((((((((........))))))))))))-))))))))............------------ ( -28.60, z-score = -2.32, R) >droGri2.scaffold_15245 4897169 97 + 18325388 --AGAAACGCACGUUGACAGAGAGCAAAUAUUUUGACAGCUGUCAGUUGAAGAAAAUGAUUUUACAUUUUUUUUUUUUUGCUGCUGACUUUUUCUUCGU------------ --........(((..((.((((..(((.....))).((((.((.((..((((((((((......))))))))))..)).)).)))).)))).))..)))------------ ( -19.60, z-score = -0.33, R) >consensus AGCCACGCAGCCAUCGACAGAGAGCAAAUAUUUUGACAGCUGUCAGUCGAGGGAAAUGAUUUCGCAUUUCUGUU_____GCUUCCACUUUUUCC_CGGGGC__________ ..((.........(((((.((.(((.............))).)).))))).(((((((......)))))))..........................))............ (-14.70 = -13.79 + -0.91)

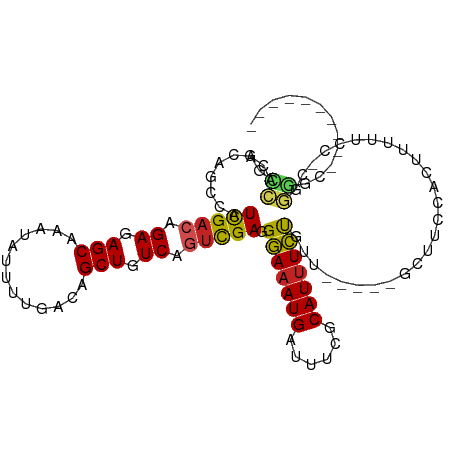
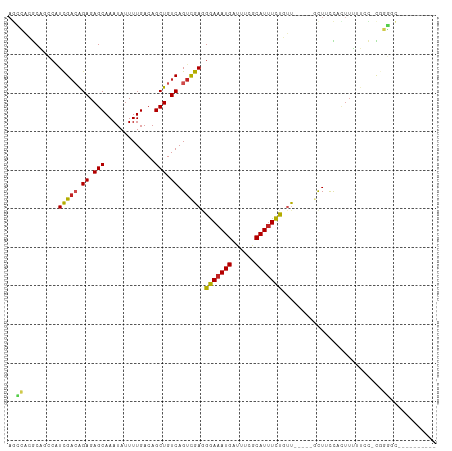
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:27:16 2011