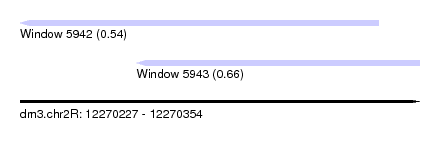
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,270,227 – 12,270,354 |
| Length | 127 |
| Max. P | 0.661244 |

| Location | 12,270,227 – 12,270,341 |
|---|---|
| Length | 114 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 57.28 |
| Shannon entropy | 0.79326 |
| G+C content | 0.61924 |
| Mean single sequence MFE | -26.26 |
| Consensus MFE | -7.91 |
| Energy contribution | -6.67 |
| Covariance contribution | -1.24 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.537218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
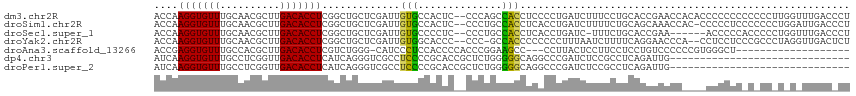
>dm3.chr2R 12270227 114 - 21146708 ACCAAGGUGUUUGCAACGCUUGACACCUCGGCUGCUCGAUUGUGCCACUC--CCCAGCCACCUCCCCUGAUCUUUCCUGCACCGAACCACACCCCCCCCCCCCUUGGUUUGACCCU ....(((((((.((...))..))))))).(((((...((.((...)).))--..))))).......................(((((((...............)))))))..... ( -22.36, z-score = -1.34, R) >droSim1.chr2R 11002143 113 - 19596830 ACCAAGGUGUUUGCAACGCUUGACACCUCGGCUGCUCGAUUGUGCCACUC--CCCUGCCACCUCACCUGAUCUUUUCUGCAGCAAACCAC-CCCCCUCCCCCCCUGGAUUGACCCU .((((((((((.((...))..)))))))..(((((..((..(((.((...--...)).))).))....((.....)).))))).......-.............)))......... ( -22.20, z-score = -1.11, R) >droSec1.super_1 9767887 107 - 14215200 ACCAAGGUGUUUGCAACGCUUGACACCUCGGCUGCUCGAUUGUGCCCCUC--CCCUGCCACCUCACCUGAUC-UUUCUGCACCGAA------ACCCCCACCCCCUGGUUUGACCCU ((((.((((((.((...))..))))))((((.(((..(((((((......--...........)))..))))-.....))))))).------............))))........ ( -19.83, z-score = -0.74, R) >droYak2.chr2R 4424129 111 + 21139217 ACCAAGGUGUUUGCAACGCUUGACACCUCGGCUGCUCGAUUGUGGCACCC--CCC-GCCACCCCCCCCUUUAAUCUUUUCAGGAACCCA--CCUCCUCCCGCCCUAGGUUGACUCU (((.(((((((.((...))..))))))).(((.........(((((....--...-)))))...................((((.....--..))))...)))...)))....... ( -25.60, z-score = -1.13, R) >droAna3.scaffold_13266 15646043 93 - 19884421 ACCGAGGUGUUUGCCACGCUUGACACCUCGUCUGGG-CAUCCCUCCACCCCACCCGGAAGCC---CCUUACUCCUUCCUCCUGUCCCCCCGUGGGCU------------------- ..(((((((((.((...))..)))))))))...(((-(.(((.............))).)))---)................((((......)))).------------------- ( -28.02, z-score = -1.80, R) >dp4.chr3 15902166 86 + 19779522 AUCAAGGUGUUUGCCUCGGUUGACACCUCAUCAGGGUCGCCUCCCCGCACCGCUCUGGGGGCAGGCCCGAUCUCCGCCUCAGAUUG------------------------------ ....(((((((.((....)).))))))).....(((((((((((..((...))...)))))).)))))(((((.......))))).------------------------------ ( -32.90, z-score = -1.16, R) >droPer1.super_2 4974469 86 + 9036312 AUCAAGGUGUUUGCCUCGGUUGACACCUCAUCAGGGUCGCCUCCCCGCACCGCUCUGGGGGCAGGCCCGAUCUCCGCCUCAGAUUG------------------------------ ....(((((((.((....)).))))))).....(((((((((((..((...))...)))))).)))))(((((.......))))).------------------------------ ( -32.90, z-score = -1.16, R) >consensus ACCAAGGUGUUUGCAACGCUUGACACCUCGGCUGCUCGAUUGUGCCACUC__CCCUGCCACCUCACCUGAUCUUUUCUUCAGCUAACC____CCCCUC_C_CC_U_G_UUGAC_CU ....(((((((..........))))))).............(((..............)))....................................................... ( -7.91 = -6.67 + -1.24)
| Location | 12,270,264 – 12,270,354 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 65.60 |
| Shannon entropy | 0.67330 |
| G+C content | 0.58708 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -9.36 |
| Energy contribution | -8.54 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.661244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
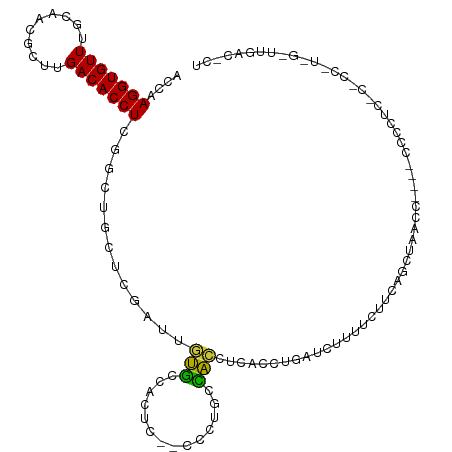
>dm3.chr2R 12270264 90 - 21146708 ---AGGGUUCGAUGUCACCAAGGUGUUUGCAACGCUUGACACCUCGGCUGCUCGAUUGUGCCACUCCC--CAGCCACCUCCCCUGAUCUUUCCUG ---((((.((((.((..((.(((((((.((...))..))))))).))..))))))..(((.(......--..).)))...))))........... ( -23.60, z-score = -0.79, R) >droSim1.chr2R 11002179 90 - 19596830 ---AGGGUUCGAUGUCACCAAGGUGUUUGCAACGCUUGACACCUCGGCUGCUCGAUUGUGCCACUCCC--CUGCCACCUCACCUGAUCUUUUCUG ---.((((((((.((..((.(((((((.((...))..))))))).))..))))))..(((.((.....--.)).)))...))))........... ( -22.80, z-score = -0.69, R) >droSec1.super_1 9767918 89 - 14215200 ---AGGGUUCGAUGUCACCAAGGUGUUUGCAACGCUUGACACCUCGGCUGCUCGAUUGUGCCCCUCCC--CUGCCACCUCACCUGAUC-UUUCUG ---.((((((((.((..((.(((((((.((...))..))))))).))..))))))....)))).....--..................-...... ( -23.00, z-score = -0.92, R) >droYak2.chr2R 4424161 93 + 21139217 --AGGGGUUCGAUGUCACCAAGGUGUUUGCAACGCUUGACACCUCGGCUGCUCGAUUGUGGCACCCCCCGCCACCCCCCCCUUUAAUCUUUUCAG --(((((.((((.((..((.(((((((.((...))..))))))).))..))))))..(((((.......)))))...)))))............. ( -33.10, z-score = -3.31, R) >droEre2.scaffold_4845 8999449 79 + 22589142 --AGGGGUUCGAUGUCACCAAGGUGUUUGCAUCGCUUGACACCUCGGCUGGUCGAUUGUGGCACCCC---CAGAUAACCCCCCU----------- --.((((((((((..((((.(((((((.((...))..))))))).)).)))))))(((.((....))---)))...)))))...----------- ( -26.80, z-score = -0.71, R) >droAna3.scaffold_13266 15646062 90 - 19884421 AGGGGUGUUCAAUGUCACCGAGGUGUUUGCCACGCUUGACACCUCGUCUGGG-CAUCCCUCCACCCCACCCGGAAGCCCCUUACUCCUUCC---- .((((((....(((((..(((((((((.((...))..)))))))))....))-))).....))))))....(((((..........)))))---- ( -34.70, z-score = -3.06, R) >dp4.chr3 15902194 74 + 19779522 AGGGGUGUUCGAUGUCAUCAAGGUGUUUGCCUCGGUUGACACCUCAUCAGGGUCGCCUCCCCGCACCGCUCUGG--------------------- ((.(((((..((((......(((((((.((....)).))))))))))).(((......))).))))).))....--------------------- ( -26.50, z-score = -0.98, R) >droPer1.super_2 4974497 74 + 9036312 AGGGGUGUUCGAUGUCAUCAAGGUGUUUGCCUCGGUUGACACCUCAUCAGGGUCGCCUCCCCGCACCGCUCUGG--------------------- ((.(((((..((((......(((((((.((....)).))))))))))).(((......))).))))).))....--------------------- ( -26.50, z-score = -0.98, R) >consensus ___GGGGUUCGAUGUCACCAAGGUGUUUGCAACGCUUGACACCUCGGCUGCUCGAUUGUGCCACCCCC__CUGCCACCCCCCCU_AUC_UU____ ....((((............(((((((..........))))))).....((......))...))))............................. ( -9.36 = -8.54 + -0.83)
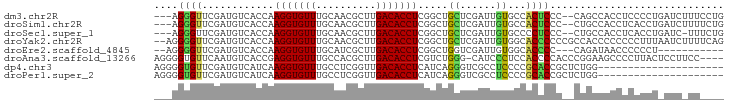

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:27:12 2011