
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,223,529 – 12,223,660 |
| Length | 131 |
| Max. P | 0.554073 |

| Location | 12,223,529 – 12,223,660 |
|---|---|
| Length | 131 |
| Sequences | 12 |
| Columns | 151 |
| Reading direction | forward |
| Mean pairwise identity | 68.85 |
| Shannon entropy | 0.63574 |
| G+C content | 0.39769 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -11.02 |
| Energy contribution | -11.37 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.554073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
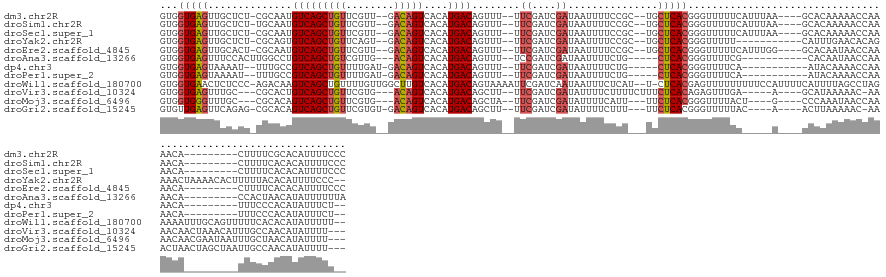
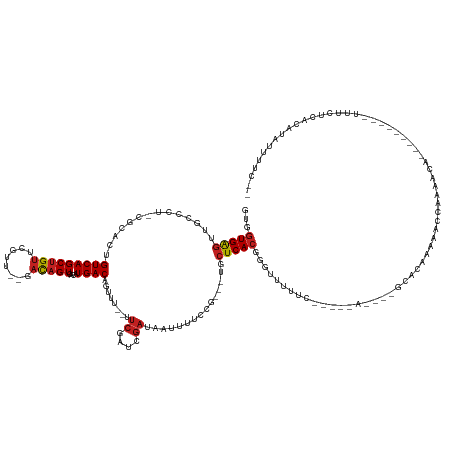
>dm3.chr2R 12223529 131 + 21146708 GUGGUGAGUUGCUCU-CGCAAUGUCAGCUGUUCGUU--GACAGUCACAUGACAGUUU--UUCGAUCGAUAAUUUUCCGC--UGCUCACGGGUUUUUCAUUUAA----GCACAAAAACCAAAACA---------CUUUUCGCACAUUUUCCC (((((((((.((..(-((...(((((((.....)))--))))(((....))).....--..)))..((......)).))--.)))))).(((((((.......----....)))))))......---------.......)))........ ( -30.50, z-score = -1.24, R) >droSim1.chr2R 10954296 131 + 19596830 GUGGUGAGUUGCUCU-UGCAAUGUCAGCUGUUCGUU--GACAGUCACAUGACAGUUU--UUCGAUCGAUAAUUUUCCGC--UGCUCACGGGUUUUUCAUUUAA----GCACAAAAACCAAAACA---------CUUUUCACACAUUUUCCC (((((((((.((...-.....(((((((.....)))--))))(((....))).....--..................))--.)))))).(((((((.......----....)))))))......---------.......)))........ ( -30.70, z-score = -1.55, R) >droSec1.super_1 9721265 131 + 14215200 GUGGUGAGUUGCUCU-CGCAAUGUCAGCUGUUCGUU--GACAGUCACAUGACAGUUU--UUCGAUCGAUAAUUUUCCGC--UGCUCACGGGUUUUUCAUUUAA----GCACAAAAACCAAAACA---------CUUUUCACACAUUUUCCC (((((((((.((..(-((...(((((((.....)))--))))(((....))).....--..)))..((......)).))--.)))))).(((((((.......----....)))))))......---------.......)))........ ( -30.80, z-score = -1.67, R) >droYak2.chr2R 4376581 131 - 21139217 GUGGUGAGUUGCUCU-CGCAGUGUCAGCUGUUCAGU--GACAGUCACAUGACAGUUU--UUCGAUCGAUAAUUUUCCGC--UGCUCACGGGUUUUU-----------CAUUUGAACACAGAAACUAAAACACUUUUUACACAUUUUCCC-- ((((((((.....))-)))(((((.(((((((..((--(.....)))..))))))).--(((((..((.((...((((.--......)))).)).)-----------)..))))).............))))).....)))........-- ( -28.60, z-score = -0.17, R) >droEre2.scaffold_4845 8950522 131 - 22589142 GUGGUGAGUUGCACU-CGCAAUGUCAGCUGUUCGUU--GACAGUCACAUGACAGUUU--UUCGAUCGAUAAUUUUCCGC--UGCUCACGGGUUUUUCAUUUGG----GCACAAUAACCAAAACA---------CUUUUCACACAUUUUCCC (((((((((.((..(-((...(((((((.....)))--))))(((....))).....--..)))..((......)).))--.))))))..........(((((----.........)))))...---------.......)))........ ( -30.80, z-score = -1.02, R) >droAna3.scaffold_13266 15599933 121 + 19884421 GUGGUGAGUUUCCACUUGGCCUGUCAGCUGUCGUUG---ACAGUCACAUGACAGUUU--UCCGAUCGAUAAUUUUCUG-----CUCACGGGUUUUCG-----------CACAAUAACCAAAACA---------CCACUAACAUAUUUUUUA (((((((((....))).(((((((.(((.((((..(---((.(((....))).))).--..)))).((......)).)-----)).)))))))....-----------..............))---------)))).............. ( -27.60, z-score = -1.11, R) >dp4.chr3 15858717 119 - 19779522 GUGGUGAGUAAAAU--UUUGCCGUCAGCUGUUUUGAU-GACAGUCACAUGACAGUUU--UUCGAUCGAUAAUUUUCUG-----CUCACGGGUUUUCA-----------AUACAAAACCAAAACA---------UUUCCCACAUAUUUCU-- ((((((((((.((.--.(((.(((((((((((.((((-....))))...))))))).--...)).)).)))..)).))-----))))).((((((..-----------....))))))......---------.....)))........-- ( -28.60, z-score = -2.28, R) >droPer1.super_2 4929737 119 - 9036312 GUGGUGAGUAAAAU--UUUGCCGUCAGCUGUUUUGAU-GACAGUCACAUGACAGUUU--UUCGAUCGAUAAUUUUCUG-----CUCACGGGUUUUCA-----------AUACAAAACCAAAACA---------UUUCCCACAUAUUUCU-- ((((((((((.((.--.(((.(((((((((((.((((-....))))...))))))).--...)).)).)))..)).))-----))))).((((((..-----------....))))))......---------.....)))........-- ( -28.60, z-score = -2.28, R) >droWil1.scaffold_180700 6042731 145 - 6630534 GUGGUGAACUCUCCC-AGACAAGUCAGCUGUUUUGUUGGCUUGUCACAUGACAGUAAAAUUCGAUCAAUAAUUUCUCAU--U-CUCACGAGUUUUUUUUUCCAUUUUCAUUUUAGCCUAGAAAAUUUGCAGUUUUUCACACAUAUUUUU-- ((((.(((((.....-.(((((((((((......)))))))))))........(((((................(((..--.-.....)))............(((((...........))))))))))))))).))))..........-- ( -29.70, z-score = -2.16, R) >droVir3.scaffold_10324 524650 130 + 1288806 GUGGUGAGUUUGC---CGCACUGUCAGCUGUUCGUG---ACAGUCACAUGACAGCUU--UUCGAUCGAUAUUUUCUUUUCUUUCUCACAGAGUUUGA-----A----GCAUAAAAAC-AAAACAACUAAACAUUUGCCAACAUAUUUU--- .(((..(((....---....((((.(((((((.(((---.....)))..))))))).--...((..((..........))..))..)))).((((..-----.----......))))-.............)))..))).........--- ( -22.10, z-score = 1.05, R) >droMoj3.scaffold_6496 2352680 129 + 26866924 GUGGUGGGUUUGC---CGCACAGUCAGCUGUUCGUG---ACAGUCACAUGACAGCUA--UUCGAUCGAUAUUUUCAUU---UUCUCACGGGUUUUACU----G----CCCAAAUAACCAAAACAACGAAUAAUUUGCUAACAUAUUUU--- (((((......))---)))..(((..((((((.(((---.....)))..))))))((--((((...((.....))...---.......((((......----)----)))...............))))))....)))..........--- ( -29.20, z-score = -1.08, R) >droGri2.scaffold_15245 3233373 132 - 18325388 GUGUUGAGUUCAGAG-CGCACAGUCAGCUGUUCGUGU-GACAGUCACAUGACAGCUU--UUCGAUCGAUAUUUUCUUU---UUCUCACGGGUUUUUAC----A----ACUUAAAAAC-AAACUAACUAGCUAAUUGCCAACAUAUUUU--- ((((((.((....((-(...(.(((((((((.(((((-(.....)))))))))))).--...))).)...........---.........(((((((.----.----...)))))))-..........)))....)))))))).....--- ( -31.60, z-score = -1.97, R) >consensus GUGGUGAGUUGCCCU_CGCACUGUCAGCUGUUCGUU__GACAGUCACAUGACAGUUU__UUCGAUCGAUAAUUUUCCG___UGCUCACGGGUUUUUC_____A____GCACAAAAACCAAAACA_________UUUCUCACAUAUUUUC__ ...(((((..............(((((((((........)))))....))))........((....))...............)))))............................................................... (-11.02 = -11.37 + 0.35)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:27:04 2011