| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,223,097 – 12,223,194 |
| Length | 97 |
| Max. P | 0.568854 |

| Location | 12,223,097 – 12,223,194 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 63.99 |
| Shannon entropy | 0.65511 |
| G+C content | 0.53220 |
| Mean single sequence MFE | -32.49 |
| Consensus MFE | -9.49 |
| Energy contribution | -9.86 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.568854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
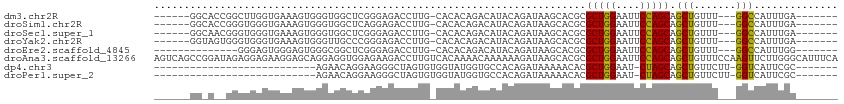
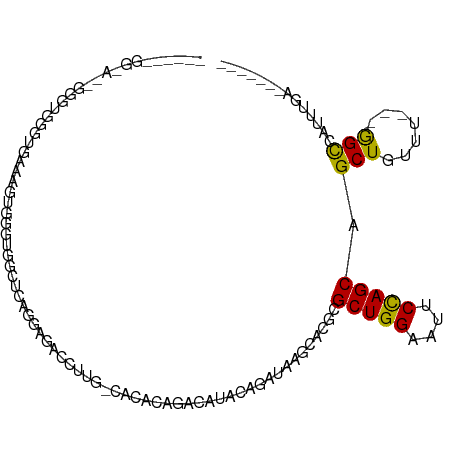
>dm3.chr2R 12223097 97 + 21146708 ------GGCACCGGCUUGGUGAAAGUGGGUGGCUCGGGAGACCUUG-CACACAGACAUACAGAUAAGCACGCGCUGGAAUUCCAGCAGCUGUUU---GGCCAUUUGA------- ------..((((.....)))).(((((((((((..((....))..)-).)))...........((((((.(((((((....))))).)))))))---).))))))..------- ( -37.00, z-score = -1.91, R) >droSim1.chr2R 10953855 97 + 19596830 ------GGCACCGGGUGGGUGAAAGUGGGUGGCUCAGGAGACCUUG-CACACAGACAUACAGAUAAGCACGCGCUGGAAUUCCAGCAGCUGUUU---GGCCAUUUGA------- ------..((((.....)))).(((((((((((...(....)...)-).)))...........((((((.(((((((....))))).)))))))---).))))))..------- ( -33.40, z-score = -0.81, R) >droSec1.super_1 9720824 97 + 14215200 ------GGCAACGGGUGGGUGAAAGUGGGUGGCUCGGGAGACCUUG-CACACAGACAUACAGAUAAGCACGCGCUGGAAUUCCAGCAGCUGUUU---GGCCAUUUGA------- ------.....(((((((......((..(((((..((....))..)-).)))..)).......((((((.(((((((....))))).)))))))---).))))))).------- ( -36.60, z-score = -2.12, R) >droYak2.chr2R 4376135 97 - 21139217 ------GGUAGUGGGUGGGUGAAAGUGGGUUGCCCGGGAGACCUUG-CACACAGACAUACAGAUAAGCACGCGCUGGAAUUCCAGCAGCUGUUU---GGCCAUUUGA------- ------(((.(((.(((((..(.......)..)))((....))..)-).)))...........((((((.(((((((....))))).)))))))---))))......------- ( -34.20, z-score = -1.09, R) >droEre2.scaffold_4845 8950092 89 - 22589142 --------------GGGAGUGGGAGUGGGCGGCUCGGGAGACCUUG-CACACAGACAUACAGAUAAGCACGCGCUGGAAUUCCAGCAGCUGUUU---GGCCAUUUGG------- --------------..((((((..(((....((..((....))..)-).)))...........((((((.(((((((....))))).)))))))---).))))))..------- ( -32.70, z-score = -1.96, R) >droAna3.scaffold_13266 15599400 114 + 19884421 AGUCAGCCGGAUAGAGGAGAAGGAGCAGGAGGUGGAGAAGACCUUGUCACAAAACAAAAAAGAUAAGCACGCGCUGGAAUUCCAGCAGCUGUUUCCAAGUUCUUGGGCAUUUCA .(((..((.......))........((((((.((((((....((((((.............))))))...(((((((....))))).))..))))))..)))))))))...... ( -30.62, z-score = -0.41, R) >dp4.chr3 15858169 78 - 19779522 ---------------------------AGAACAGGAAGGGCUAGUGUGGUAUGGUGCCACAGAUAAAAACACGCUGGAAU-CUAGCAGCUGUUCUU-GGUCAUUCGC------- ---------------------------.((.((((((.((((..((((((.....))))))...........(((((...-))))))))).)))))-).))......------- ( -27.70, z-score = -2.91, R) >droPer1.super_2 4929195 78 - 9036312 ---------------------------AGAACAGGAAGGGCUAGUGUGGUAUGGUGCCACAGAUAAAAACACGCUGGAAU-CUAGCAGCUGUUCUU-GGUCAUUCGC------- ---------------------------.((.((((((.((((..((((((.....))))))...........(((((...-))))))))).)))))-).))......------- ( -27.70, z-score = -2.91, R) >consensus ______GG_A__GGGUGGGUGAAAGUGGGUGGCUCAGGAGACCUUG_CACACAGACAUACAGAUAAGCACGCGCUGGAAUUCCAGCAGCUGUUU___GGCCAUUUGA_______ ................................................................(((((.(((((((....))))).))))))).................... ( -9.49 = -9.86 + 0.37)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:27:03 2011