| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,221,178 – 12,221,273 |
| Length | 95 |
| Max. P | 0.999193 |

| Location | 12,221,178 – 12,221,273 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 78.50 |
| Shannon entropy | 0.38227 |
| G+C content | 0.49774 |
| Mean single sequence MFE | -30.83 |
| Consensus MFE | -19.09 |
| Energy contribution | -19.49 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
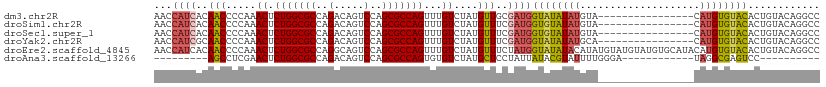
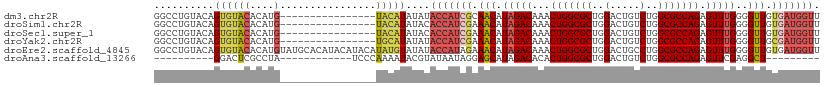
>dm3.chr2R 12221178 95 + 21146708 AACCAUCACAACCCCAAACUCUGGCGCCAGACAGUCCAGCGCCAGUUUGUCUAUGUUGCGAUGGUAUAUAUGUA----------------CAUGUGUACACUGUACAGGCC .((((((.((((.....((.(((((((..(.....)..)))))))...))....)))).)))))).....((((----------------((.((....)))))))).... ( -33.20, z-score = -3.06, R) >droSim1.chr2R 10951955 95 + 19596830 AACCAUCACAACCCCAAACUCUGGCGCCAGACAGUCCAGCGCCAGUUUGUCUAUGUUUCGAUGGUGUAUAUGUA----------------CAUGUGUACACUGUACAGGCC .(((((((((....((((((..(((((..(.....)..)))))))))))....)))...))))))(((((((((----------------(....))))).)))))..... ( -30.90, z-score = -2.52, R) >droSec1.super_1 9718935 95 + 14215200 AACCAUCACAACCCCAAACUCUGGCGCCAGACAGUCCAGCGCCAGUUUGUCUAUGUUUCGAUGGUGUAUAUGUA----------------CAUGUGUACACUGUACAGGCC .(((((((((....((((((..(((((..(.....)..)))))))))))....)))...))))))(((((((((----------------(....))))).)))))..... ( -30.90, z-score = -2.52, R) >droYak2.chr2R 4374179 95 - 21139217 AACCAUCGCAACCCCAAACUCUGGCGCCAGACAGUCCAGCGCCAGUUUGUCUAUGUUUCGAUGGUAUAUAUGCA----------------CAUGUGUACACUGUACAGGCC .(((((((.(((.....((.(((((((..(.....)..)))))))...))....))).))))))).........----------------..(.(((((...))))).).. ( -28.20, z-score = -1.80, R) >droEre2.scaffold_4845 8948184 111 - 22589142 AACCAUCACAACCCCAAACUCUGGCGCCAGGCAGUCCAGCGCCAGUUUGUCUAUGUUUCUAUGGUAUAUACAUAUGUAUGUAUGUGCAUACAUGUGUACACUGUACAGGCC ..............((((((..(((((..((....)).))))))))))).....((((.((((((...(((((((((((((....))))))))))))).)))))).)))). ( -35.00, z-score = -1.81, R) >droAna3.scaffold_13266 15597900 80 + 19884421 ---------AGCCUCGAACUCUGGCGCCAGACAGUCCAGCGCCAGUGUGUCUAUGCUCCUAUUAUACGUAUUUUGGGA------------UAGGCGAGUCC---------- ---------...(((..((.(((((((..(.....)..))))))).))(((((...(((((..((....))..)))))------------))))))))...---------- ( -26.80, z-score = -1.66, R) >consensus AACCAUCACAACCCCAAACUCUGGCGCCAGACAGUCCAGCGCCAGUUUGUCUAUGUUUCGAUGGUAUAUAUGUA________________CAUGUGUACACUGUACAGGCC ...((((..(((.....((.(((((((..(.....)..)))))))...))....)))..))))(((((((((..................)))))))))............ (-19.09 = -19.49 + 0.39)
| Location | 12,221,178 – 12,221,273 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 78.50 |
| Shannon entropy | 0.38227 |
| G+C content | 0.49774 |
| Mean single sequence MFE | -38.85 |
| Consensus MFE | -28.55 |
| Energy contribution | -29.67 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.50 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.70 |
| SVM RNA-class probability | 0.999193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
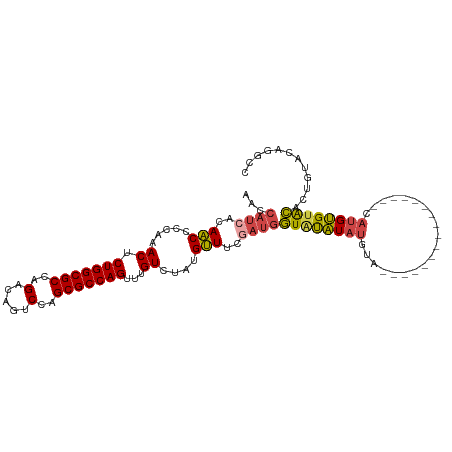
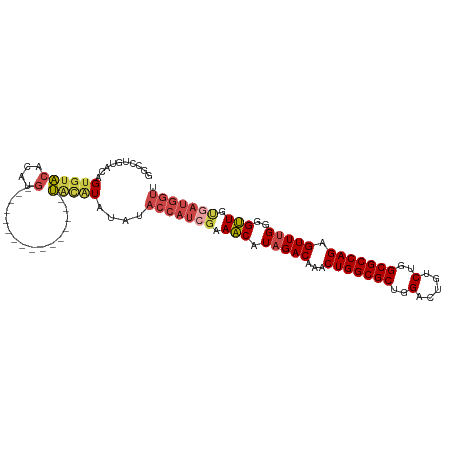
>dm3.chr2R 12221178 95 - 21146708 GGCCUGUACAGUGUACACAUG----------------UACAUAUAUACCAUCGCAACAUAGACAAACUGGCGCUGGACUGUCUGGCGCCAGAGUUUGGGGUUGUGAUGGUU ....((((((((....)).))----------------)))).....(((((((((((.(((((...(((((((..(.....)..))))))).)))))..))))))))))). ( -44.80, z-score = -5.03, R) >droSim1.chr2R 10951955 95 - 19596830 GGCCUGUACAGUGUACACAUG----------------UACAUAUACACCAUCGAAACAUAGACAAACUGGCGCUGGACUGUCUGGCGCCAGAGUUUGGGGUUGUGAUGGUU .....(((..((((((....)----------------))))).)))(((((((.(((.(((((...(((((((..(.....)..))))))).)))))..))).))))))). ( -39.40, z-score = -3.67, R) >droSec1.super_1 9718935 95 - 14215200 GGCCUGUACAGUGUACACAUG----------------UACAUAUACACCAUCGAAACAUAGACAAACUGGCGCUGGACUGUCUGGCGCCAGAGUUUGGGGUUGUGAUGGUU .....(((..((((((....)----------------))))).)))(((((((.(((.(((((...(((((((..(.....)..))))))).)))))..))).))))))). ( -39.40, z-score = -3.67, R) >droYak2.chr2R 4374179 95 + 21139217 GGCCUGUACAGUGUACACAUG----------------UGCAUAUAUACCAUCGAAACAUAGACAAACUGGCGCUGGACUGUCUGGCGCCAGAGUUUGGGGUUGCGAUGGUU ....((((((((....)).))----------------)))).....(((((((.(((.(((((...(((((((..(.....)..))))))).)))))..))).))))))). ( -40.10, z-score = -3.82, R) >droEre2.scaffold_4845 8948184 111 + 22589142 GGCCUGUACAGUGUACACAUGUAUGCACAUACAUACAUAUGUAUAUACCAUAGAAACAUAGACAAACUGGCGCUGGACUGCCUGGCGCCAGAGUUUGGGGUUGUGAUGGUU ..........(((((((.(((((((......))))))).)))))))(((((...(((.(((((...(((((((..(.....)..))))))).)))))..)))...))))). ( -42.20, z-score = -2.90, R) >droAna3.scaffold_13266 15597900 80 - 19884421 ----------GGACUCGCCUA------------UCCCAAAAUACGUAUAAUAGGAGCAUAGACACACUGGCGCUGGACUGUCUGGCGCCAGAGUUCGAGGCU--------- ----------......((((.------------(((................))).....((.((.(((((((..(.....)..))))))).)))).)))).--------- ( -27.19, z-score = -1.92, R) >consensus GGCCUGUACAGUGUACACAUG________________UACAUAUAUACCAUCGAAACAUAGACAAACUGGCGCUGGACUGUCUGGCGCCAGAGUUUGGGGUUGUGAUGGUU ..............................................(((((((.(((.(((((...(((((((..(.....)..))))))).)))))..))).))))))). (-28.55 = -29.67 + 1.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:27:00 2011