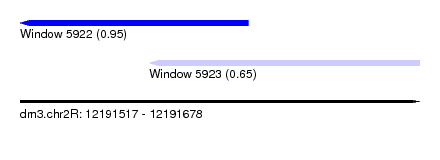
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,191,517 – 12,191,678 |
| Length | 161 |
| Max. P | 0.952336 |

| Location | 12,191,517 – 12,191,609 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 80.05 |
| Shannon entropy | 0.37457 |
| G+C content | 0.56724 |
| Mean single sequence MFE | -37.15 |
| Consensus MFE | -19.71 |
| Energy contribution | -21.06 |
| Covariance contribution | 1.35 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
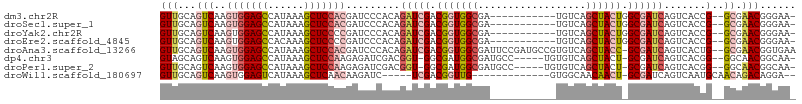
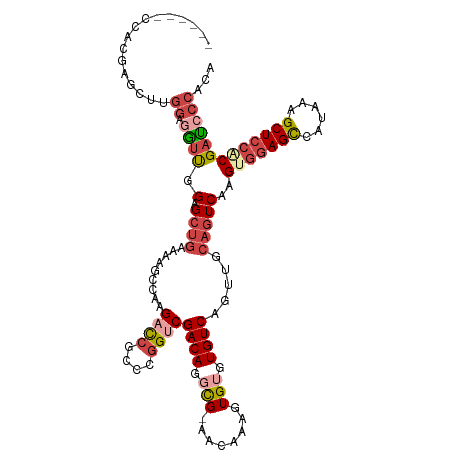
>dm3.chr2R 12191517 92 - 21146708 GUUGCAGUCAAGUGGAGCCAUAAAGCUCCACGAUCCCACAGAUCGACGGUGGCGA-----------UGUCAGCUACUGGCGAUCAGUCACCG--GCGAACGGGAA- .((((.(((..(((((((......)))))))((((.....)))))))((((((((-----------((((((...))))).))).)))))).--)))).......- ( -40.40, z-score = -3.69, R) >droSec1.super_1 9690789 92 - 14215200 GUUGCAGUCAAGUGGAGCCAUAAAGCUCCACGAUCCCACAGAUCGACGGUGGCGA-----------UGUCAGCUACUGGCGAUCAGUCACCG--GCGAACGGGAA- .((((.(((..(((((((......)))))))((((.....)))))))((((((((-----------((((((...))))).))).)))))).--)))).......- ( -40.40, z-score = -3.69, R) >droYak2.chr2R 4344298 92 + 21139217 GUUGCAGUCAAGUGGAGCCAUAAAGCUCCCCGAUCCCACAGAUCGACGGUGGCGA-----------UGUCAGCUACUGGCGAUCAGUCACCG--GCGAACGGGAA- .((((.(((..(.(((((......))))).)((((.....)))))))((((((((-----------((((((...))))).))).)))))).--)))).......- ( -35.60, z-score = -2.03, R) >droEre2.scaffold_4845 8918448 92 + 22589142 GUUGCAGUCAAGUGGAGCCACAAAGCUCCCCGAUCCCACAGAUCGACGGUGGCGA-----------UGUCAGCUACUGGCGAUCAGUCACCG--GCGAACGGGAA- .((((.(((..(.(((((......))))).)((((.....)))))))((((((((-----------((((((...))))).))).)))))).--)))).......- ( -35.60, z-score = -1.84, R) >droAna3.scaffold_13266 15569787 103 - 19884421 GUUGCAGUCAAGUGGAGCCAUAAAGCUCCACGAUCCCACAGAUCGACGGUGGCGAUUCCGAUGCCGUGUCAGCUACC-GCGAUCAGUCACUG--GCGAACGGUGAA (((((.(((..(((((((......)))))))((((.....)))))))(((((((((..((....)).))).))))))-)))))...((((((--.....)))))). ( -44.40, z-score = -3.66, R) >dp4.chr3 15820782 96 + 19779522 GUAGCAGUCAAGUGGAGCCAUAAAGCUCCAAGAGAUCGACGGU-GGCGAUGGCGAUGCC-----UGUGUCAGCUACU-GCGAUCAGUCACGG--GGCAACGGCAA- ...((.(((..(((((((......)))))....(((((.((((-(((((((((...)))-----...))).))))))-))))))....))..--)))....))..- ( -36.10, z-score = -1.63, R) >droPer1.super_2 4894337 96 + 9036312 GUUGCAGUCAAGUGGAGCCAUAAAGCUCCAAGAGAUCGACGGU-GGCGAUGGCGAUGCC-----UGUGUCAGCUACU-GCGAUCAGUCACGG--GGCAACGGCAA- (((((..((...((((((......))))))...(((((.((((-(((((((((...)))-----...))).))))))-))))))......))--.))))).....- ( -38.70, z-score = -2.18, R) >droWil1.scaffold_180697 1764470 85 - 4168966 GUUGCAGUCAAGUGGAGUCAUAAAGCUCAACAAGAUC-----UCGACGGUUG-------------GUGGCAACAACU-GCGAUCAGUCAAUGCAACAGACAGGA-- (((((((((..((.((((......)))).))..(...-----.))))(..((-------------((.(((.....)-)).))))..)..))))))........-- ( -26.00, z-score = -1.96, R) >consensus GUUGCAGUCAAGUGGAGCCAUAAAGCUCCACGAUCCCACAGAUCGACGGUGGCGA___________UGUCAGCUACU_GCGAUCAGUCACCG__GCGAACGGGAA_ .((((......(((((((......))))))).........(((((.(((((((..................)))))).))))))..........))))........ (-19.71 = -21.06 + 1.35)
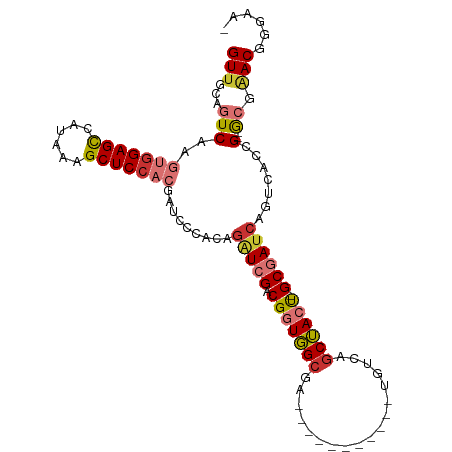
| Location | 12,191,569 – 12,191,678 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 78.30 |
| Shannon entropy | 0.39705 |
| G+C content | 0.55843 |
| Mean single sequence MFE | -36.65 |
| Consensus MFE | -20.81 |
| Energy contribution | -22.65 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.654117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12191569 109 - 21146708 ------CCACAAGCUUGGAGGUUGGAGCUGAAAAGCCAAGACCGCCCGGUCGACAGGCG-AACAAAGUGUGUGUCAGUUGCAGUCAAGUGGAGCCAUAAAGCUCCACGAUCCCACA ------..........((((..((.((((((...(((..((((....))))....))).-.(((.....))).)))))).))..)..(((((((......)))))))..))).... ( -38.70, z-score = -2.30, R) >droSec1.super_1 9690841 112 - 14215200 ---GGACCACGAGCUUGGAGGUUGGAGCUGAAAAGCCAAGACCGCCCGGUCGACAGGCG-AACAAAGUGUGUGUCAGUUGCAGUCAAGUGGAGCCAUAAAGCUCCACGAUCCCACA ---((((((......))).(..((.((((((...(((..((((....))))....))).-.(((.....))).)))))).))..)..(((((((......)))))))..))).... ( -39.80, z-score = -1.89, R) >droYak2.chr2R 4344350 115 + 21139217 GUUGGACCACGAGCUUGGAGGUUGGAGCUGAAAACCCAAGACCGCCCGGUCGACAGGCG-AACAAAGUGUGUGUCAGUUGCAGUCAAGUGGAGCCAUAAAGCUCCCCGAUCCCACA ((.(((.(....((((((...(((.((((((........((((....)))).(((.((.-......)).))).)))))).)))))))))(((((......)))))..).))).)). ( -37.40, z-score = -1.00, R) >droEre2.scaffold_4845 8918500 112 + 22589142 ---GGACCACGAGCUUGGAGGUUCGAGCUGAAAGCCCAAGACCGCCCGGUCGACAGGCG-AACAAAGUGUGUGUCAGUUGCAGUCAAGUGGAGCCACAAAGCUCCCCGAUCCCACA ---(((.(..((((((.(.(((((..(((((..((....((((....))))((((.(((-.......))).))))....))..)).))).))))).).))))))...).))).... ( -37.40, z-score = -1.13, R) >droAna3.scaffold_13266 15569850 99 - 19884421 ---------------CGGGAAUUUG-GCUGAAAUGUGGAGACCGCCCGGGCGACAGCUG-AACAAAGUGUGUGUCAGUUGCAGUCAAGUGGAGCCAUAAAGCUCCACGAUCCCACA ---------------.((((..(((-((((......(....)(((....))).((((((-((((.....))).))))))))))))))(((((((......)))))))..))))... ( -41.90, z-score = -3.72, R) >droMoj3.scaffold_6496 1694059 93 - 26866924 ------------------CAGACAGAGACAGACAGCACGGAUAGCAGGGGCGACAGCUGAAACAAAGUGUGUGUCAGUUGCAGUCAAGUGGAGUCAUAAAGCU--GCGGCCCA--- ------------------................((.......))..((((...((((((.(((.....))).))))))(((((...(((....)))...)))--)).)))).--- ( -24.70, z-score = -0.01, R) >consensus ______CCACGAGCUUGGAGGUUGGAGCUGAAAAGCCAAGACCGCCCGGUCGACAGGCG_AACAAAGUGUGUGUCAGUUGCAGUCAAGUGGAGCCAUAAAGCUCCACGAUCCCACA ................((.((((...((((.........((((....))))((((.(((........))).)))).....))))...(((((((......)))))))))))))... (-20.81 = -22.65 + 1.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:56 2011