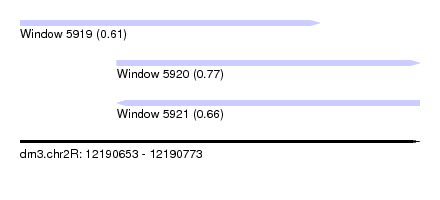
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,190,653 – 12,190,773 |
| Length | 120 |
| Max. P | 0.768967 |

| Location | 12,190,653 – 12,190,743 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 63.21 |
| Shannon entropy | 0.64655 |
| G+C content | 0.45059 |
| Mean single sequence MFE | -20.17 |
| Consensus MFE | -9.01 |
| Energy contribution | -9.12 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.605212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
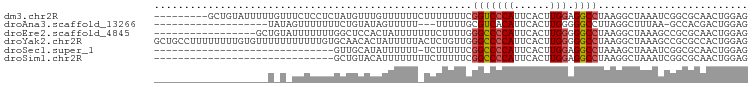
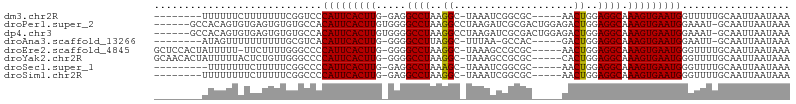
>dm3.chr2R 12190653 90 + 21146708 ---------GCUGUAUUUUUGUUUCUCCUCUAUGUUUGUUUUUUCUUUUUUUCGGUCCCAUUCACUUGGAGGCCUAAGGCUAAAUCGGCGCAACUGGAG ---------...............((((....(((..((((..((((......(((((((......))).)))).))))..))))..))).....)))) ( -17.50, z-score = -0.38, R) >droAna3.scaffold_13266 15568927 76 + 19884421 -------------------UAUAGUUUUUUUCUGUAUAGUUUUU---UUUUUGCGUCACAUUCACUUGGGGGCCUUAGGCUUUAA-GCCACGACUGGAG -------------------(((((.......)))))(((((...---.....((.((.((......)))).))....(((.....-)))..)))))... ( -12.70, z-score = 0.21, R) >droEre2.scaffold_4845 8917616 82 - 22589142 -----------------GCUGUAUUUUUUUGGCUCCACUAUUUUUUUCUUUUGGGCCCCAUUCACUUGGGGGCCUAAGGCUAAAGCCGCGCAACUGGAG -----------------((((........))))((((.............(((((((((.........)))))))))(((....))).......)))). ( -26.40, z-score = -1.46, R) >droYak2.chr2R 4343406 99 - 21139217 GCUGCCUUUUUUUUGUGUUUUUUUUUUUGUGCAACACUAUUUUUACUCUGUUGGGCCCCAUUCACUUGGGGGCCUAAGGCUAAAGCCGCGCCACUGGAG ..........((..(((...........((((..................(((((((((.........)))))))))(((....))))))))))..)). ( -26.40, z-score = -0.41, R) >droSec1.super_1 9689943 68 + 14215200 ------------------------------GUUGCAUAUUUUUU-UCUUUUUCGGCCCCAUUCACUUGGAGGCCUAAAGCUAAAUCGGCGCAACUGGAG ------------------------------(((((.........-........(((((((......))).))))....(((.....))))))))..... ( -21.40, z-score = -2.79, R) >droSim1.chr2R 10923085 69 + 19596830 ------------------------------GCUGUACAUUUUUUUUCUUUUUCGGCCCCAUUCACUUGGAGGCCUAAGGCUAAAUCGGCGCAACUGGAG ------------------------------((((......(((..((((....(((((((......))).)))).))))..))).)))).......... ( -16.60, z-score = -0.67, R) >consensus ___________________U_U_UUUUUUUGCUGCAUAUUUUUU_UUUUUUUCGGCCCCAUUCACUUGGAGGCCUAAGGCUAAAUCGGCGCAACUGGAG .....................................................(((((((......))).))))......................... ( -9.01 = -9.12 + 0.11)
| Location | 12,190,682 – 12,190,773 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 76.25 |
| Shannon entropy | 0.44610 |
| G+C content | 0.44015 |
| Mean single sequence MFE | -28.38 |
| Consensus MFE | -17.13 |
| Energy contribution | -17.03 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.768967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
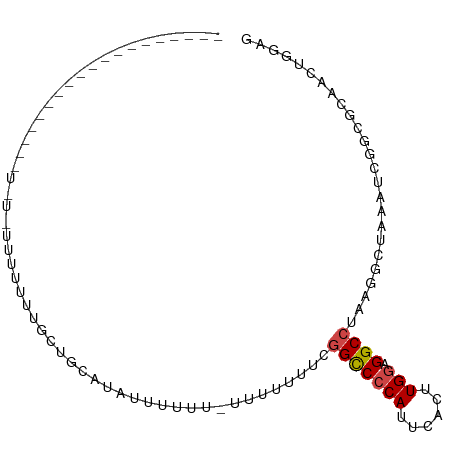
>dm3.chr2R 12190682 91 + 21146708 --------UUUUUUCUUUUUUUCGGUCCCAUUCACUUG-GAGGCCUAAGGC-UAAAUCGGCGC-----AACUGGAGGCAAAGUGAAUGGUUUUUGCAAUUAAUAAA --------...................((((((((((.-...((((.((((-(.....)))..-----..))..)))).))))))))))................. ( -21.80, z-score = -0.54, R) >droPer1.super_2 4893359 99 - 9036312 ------GCCACAGUGUGAGUGUGUGCCACAUUCACUUGUGGGGCCUAAGGCCUAAGAUCGCGACUGGAGACUGGAGGCAAAGUGAAUGGAAAU-GCAAUUAAUAAA ------(((.(((((((((((((...)))))))))((((((((((...))))).....)))))......))))..)))...............-............ ( -29.20, z-score = -1.04, R) >dp4.chr3 15819835 99 - 19779522 ------GCCACAGUGUGAGUGUGUGCCACAUUCACUUGUGGGGCCUAAGGCCUAAGAUCGCGACUGGAGACUGGAGGCAAAGUGAAUGGAAAU-GCAAUUAAUAAA ------(((.(((((((((((((...)))))))))((((((((((...))))).....)))))......))))..)))...............-............ ( -29.20, z-score = -1.04, R) >droAna3.scaffold_13266 15568943 89 + 19884421 --------AUAGUUUUUUUUUUGCGUCACAUUCACUUG-GGGGCCUUAGGC-UUUAA-GCCAC-----GACUGGAGGCAAAGUGAAUGGAAUU-GCAAUUAAUAAA --------........((..(((((...(((((((((.-...(((((.(((-.....-))).(-----....)))))).)))))))))....)-))))..)).... ( -26.70, z-score = -2.74, R) >droEre2.scaffold_4845 8917630 98 - 22589142 GCUCCACUAUUUUU-UUCUUUUGGGCCCCAUUCACUUG-GGGGCCUAAGGC-UAAAGCCGCGC-----AACUGGAGGCAAAGUGAAUGGGUUUUGCAAUUAAUAAA .(((((........-.....(((((((((.........-)))))))))(((-....)))....-----...)))))((((((........)))))).......... ( -32.10, z-score = -1.25, R) >droYak2.chr2R 4343436 99 - 21139217 GCAACACUAUUUUUACUCUGUUGGGCCCCAUUCACUUG-GGGGCCUAAGGC-UAAAGCCGCGC-----CACUGGAGGCAAAGUGAAUGGGUUUUGCAAUUAAUAAA ((((.(((...((((((...(((((((((.........-)))))))))(((-....)))..((-----(......)))..))))))..))).)))).......... ( -36.80, z-score = -2.54, R) >droSec1.super_1 9689951 90 + 14215200 ---------UUUUUUUCUUUUUCGGCCCCAUUCACUUG-GAGGCCUAAAGC-UAAAUCGGCGC-----AACUGGAGGCAAAGUGAAUGGGUUUUGCAAUUAAUAAA ---------...............(((((((((((((.-...((((...((-(.....)))..-----......)))).)))))))))))....)).......... ( -25.50, z-score = -1.31, R) >droSim1.chr2R 10923093 91 + 19596830 --------UUUUUUUUCUUUUUCGGCCCCAUUCACUUG-GAGGCCUAAGGC-UAAAUCGGCGC-----AACUGGAGGCAAAGUGAAUGGGUUUUGCAAUUAAUAAA --------................(((((((((((((.-...((((.((((-(.....)))..-----..))..)))).)))))))))))....)).......... ( -25.70, z-score = -1.12, R) >consensus ________AUUUUUUUUUUUUUGGGCCCCAUUCACUUG_GGGGCCUAAGGC_UAAAACCGCGC_____AACUGGAGGCAAAGUGAAUGGGUUUUGCAAUUAAUAAA ............................(((((((((.....((((..((....................))..)))).))))))))).................. (-17.13 = -17.03 + -0.11)


| Location | 12,190,682 – 12,190,773 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 76.25 |
| Shannon entropy | 0.44610 |
| G+C content | 0.44015 |
| Mean single sequence MFE | -23.57 |
| Consensus MFE | -13.44 |
| Energy contribution | -13.28 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.664087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12190682 91 - 21146708 UUUAUUAAUUGCAAAAACCAUUCACUUUGCCUCCAGUU-----GCGCCGAUUUA-GCCUUAGGCCUC-CAAGUGAAUGGGACCGAAAAAAAGAAAAAA-------- .................((((((((((.((((...(((-----(........))-))...))))...-.))))))))))...................-------- ( -19.70, z-score = -1.42, R) >droPer1.super_2 4893359 99 + 9036312 UUUAUUAAUUGC-AUUUCCAUUCACUUUGCCUCCAGUCUCCAGUCGCGAUCUUAGGCCUUAGGCCCCACAAGUGAAUGUGGCACACACUCACACUGUGGC------ .........(((-.....(((((((((.((((...((((..((.......)).))))...)))).....)))))))))..)))((((.......))))..------ ( -20.90, z-score = -0.26, R) >dp4.chr3 15819835 99 + 19779522 UUUAUUAAUUGC-AUUUCCAUUCACUUUGCCUCCAGUCUCCAGUCGCGAUCUUAGGCCUUAGGCCCCACAAGUGAAUGUGGCACACACUCACACUGUGGC------ .........(((-.....(((((((((.((((...((((..((.......)).))))...)))).....)))))))))..)))((((.......))))..------ ( -20.90, z-score = -0.26, R) >droAna3.scaffold_13266 15568943 89 - 19884421 UUUAUUAAUUGC-AAUUCCAUUCACUUUGCCUCCAGUC-----GUGGC-UUAAA-GCCUAAGGCCCC-CAAGUGAAUGUGACGCAAAAAAAAAACUAU-------- ........((((-...(((((((((((.((((......-----..(((-.....-)))..))))...-.))))))))).)).))))............-------- ( -21.40, z-score = -3.10, R) >droEre2.scaffold_4845 8917630 98 + 22589142 UUUAUUAAUUGCAAAACCCAUUCACUUUGCCUCCAGUU-----GCGCGGCUUUA-GCCUUAGGCCCC-CAAGUGAAUGGGGCCCAAAAGAA-AAAAAUAGUGGAGC ................(((((((((((.((((...(..-----...)(((....-)))..))))...-.)))))))))))(((((..(...-.....)..))).)) ( -27.30, z-score = -0.92, R) >droYak2.chr2R 4343436 99 + 21139217 UUUAUUAAUUGCAAAACCCAUUCACUUUGCCUCCAGUG-----GCGCGGCUUUA-GCCUUAGGCCCC-CAAGUGAAUGGGGCCCAACAGAGUAAAAAUAGUGUUGC ................(((((((((((.(((......)-----))..(((((..-.....)))))..-.)))))))))))...(((((..((....))..))))). ( -30.10, z-score = -1.47, R) >droSec1.super_1 9689951 90 - 14215200 UUUAUUAAUUGCAAAACCCAUUCACUUUGCCUCCAGUU-----GCGCCGAUUUA-GCUUUAGGCCUC-CAAGUGAAUGGGGCCGAAAAAGAAAAAAA--------- ................(((((((((((.((((..((((-----(........))-)))..))))...-.))))))))))).................--------- ( -25.30, z-score = -2.36, R) >droSim1.chr2R 10923093 91 - 19596830 UUUAUUAAUUGCAAAACCCAUUCACUUUGCCUCCAGUU-----GCGCCGAUUUA-GCCUUAGGCCUC-CAAGUGAAUGGGGCCGAAAAAGAAAAAAAA-------- ................(((((((((((.((((...(((-----(........))-))...))))...-.)))))))))))..................-------- ( -23.00, z-score = -1.67, R) >consensus UUUAUUAAUUGCAAAACCCAUUCACUUUGCCUCCAGUU_____GCGCCGAUUUA_GCCUUAGGCCCC_CAAGUGAAUGGGGCCCAAAAAAAAAAAAAU________ ................(((((((((((.((((...((..................))...)))).....))))))))).))......................... (-13.44 = -13.28 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:54 2011