| Sequence ID | dm3.chr2R |
|---|---|
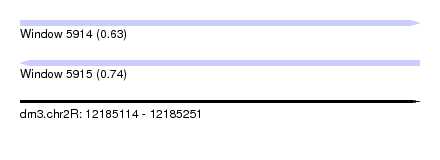
| Location | 12,185,114 – 12,185,251 |
| Length | 137 |
| Max. P | 0.744830 |

| Location | 12,185,114 – 12,185,251 |
|---|---|
| Length | 137 |
| Sequences | 12 |
| Columns | 155 |
| Reading direction | forward |
| Mean pairwise identity | 70.52 |
| Shannon entropy | 0.61454 |
| G+C content | 0.58083 |
| Mean single sequence MFE | -51.21 |
| Consensus MFE | -24.52 |
| Energy contribution | -24.74 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.630918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
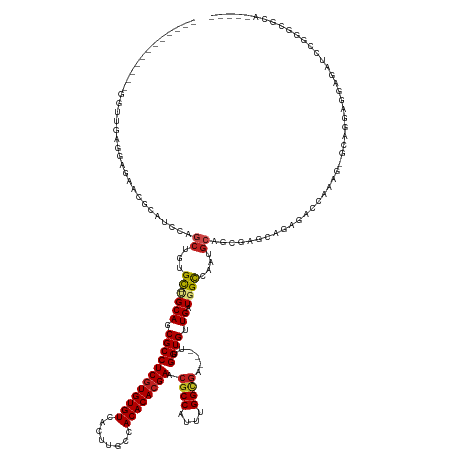
>dm3.chr2R 12185114 137 + 21146708 ------------GGCUGAGGAGAACGCAUCCAGCUGUGC-UGCAGCGCCUCGUGUGUCACUUGCCACACACGAACGCCAUUUGGCGA---UUGGUGUUGAUGGCCAAUGCGGCGAGCAGAGACCAAAG-ACUGGAGGAGAUCGGGGCGCACAGA- ------------(((((.(..(....)..))))))...(-((..((((((((.((.((.((((((((((.(((.((((....)))).---))))))).....((....))))))))......(((...-..)))..)).)))))))))).))).- ( -51.30, z-score = -0.32, R) >droSim1.chr2R 10917755 137 + 19596830 ------------GGCUGUGGAGAACGCAUCCAGCUGUGC-UGCAGCGCCUCGUGUGUCACUUGCCACACACGAACGCCAUUUGGCGA---UUGGUGUUGAUGGCCAAUGCGGCGAGCAGAGACCAAAU-ACUGGAGGAGAUCGGGGCGCACAGA- ------------((.((((.....)))).))..((((((-..(((((((((((((((........)))))))).((((....)))).---..)))))))...(((..(((.....)))....(((...-..)))..........))))))))).- ( -54.60, z-score = -1.05, R) >droSec1.super_1 9684486 137 + 14215200 ------------GGCUGUGGAGAACGCAUCCAGCUGUGC-UGCAGCGCCUCGUGUGUCACUUGCCACACACGAACGCCAUUUGGCGA---UUGGUGUUGAUGGCCAAUGGGGCGAGCAGAGACCAAAG-ACUGGAGGAGAUCGGGGCGCACAGA- ------------((.((((.....)))).))..((((((-..(((((((((((((((........)))))))).((((....)))).---..)))))))...(((..(((..(.....)...)))...-.((((......))))))))))))).- ( -53.70, z-score = -0.82, R) >droYak2.chr2R 4338155 137 - 21139217 ------------GGAUGGGGAGAACGCAUCCAGCUGUGC-UGCAGCGCCUCGUGUGUCACUUGCCACACACGAACGCCAUUUGGCGA---UUGGUGUUGAUGGCCAAUGCGGCGAGCAGAGUCCAAAG-GCUGGAGGAGAUCGGGACGCACAGU- ------------(((((.(.....).))))).(((((((-(((((((((((((((((........)))))))).((((....)))).---..))))))....(((.....)))..)))..((((...(-(((.....)).)).)))))))))))- ( -59.20, z-score = -2.15, R) >droEre2.scaffold_4845 8912379 137 - 22589142 ------------GGAUGAGGAGAACGCAUCCAGCUGUGC-CGCAGCGCCUCGUGUGUCACUUGCCACACACGAACGCCAUUUGGCGA---UUGGUGUUGAUGGCCAAUGCGGUGAGCAGAGUCCAAAG-GCUGGAGGAGAUCGGGACGCACAGA- ------------(((((.(.....).)))))..((((((-(((((((((((((((((........)))))))).((((....)))).---..))))))).)).....(((.....)))..((((...(-(((.....)).)).)))))))))).- ( -54.30, z-score = -1.30, R) >droAna3.scaffold_13266 15564081 124 + 19884421 ------------CGGCGAGGAGAACGCAUCCAGCUGUGU-UGCAGCGCCUCGUGUGUCACUUGCCACACACGAACGCCAUUUGGCGA---AUGGUGUUGAUGGCCAAUGCAGCGAUCCGAGGUUGGGG-GGACGGCAAGGA-------------- ------------......(((.......))).(((((((-(((((((((((((((((........)))))))).((((....)))).---..))))))).....)))))))))..(((...((((...-...))))..)))-------------- ( -48.10, z-score = -0.40, R) >dp4.chr3 15815486 135 - 19779522 ----------GGAGUGGACGACGACGCUUCCAGCUGUGC-UGCAUCGCCUCGUGUGUCACUUGCCACACACGAACGCCAUUUGGCGA---CUGGUGUUGAUGGCAUAUGCAGAGAACUGAGAACAGGA-GAAGCAGGCAAUUCAGGCGAA----- ----------((((((........))))))((((...))-))..((((((....((((...((((((((((.(.((((....)))).---.).))).)).)))))..(((......(((....)))..-...)))))))....)))))).----- ( -45.50, z-score = -0.19, R) >droPer1.super_2 4888340 135 - 9036312 ----------GGAGUGGACGACGACGCUUCCAGCUGUGC-UGCAUCGCCUCGUGUGUCACUUGCCACACACGAACGCCAUUUGGCGA---CUGGUGUUGAUGGCAUAUGCAGAGAACUGAGAACAGGA-GAAGCAGGCAAUUCAGGCGAA----- ----------((((((........))))))((((...))-))..((((((....((((...((((((((((.(.((((....)))).---.).))).)).)))))..(((......(((....)))..-...)))))))....)))))).----- ( -45.50, z-score = -0.19, R) >droWil1.scaffold_180697 1752621 126 + 4168966 ---------------------CGUUGAUUCCAGCUGUGCUGGCAUCGCCUCAUGUGUCACUUGCCACACACGAUAGCCAUUUGGUGCGAUUUGGUGUUGAUGGUA---GCAAGAAGUAAGGAGAAACCCGCACUGAUAUAUACCGAUGCC----- ---------------------.((((....))))......(((((((....(((((((.(((((.((.(((((((.(((..(......)..)))))))).)))).---))))).(((..((......))..))))))))))..)))))))----- ( -37.00, z-score = -0.41, R) >droVir3.scaffold_12823 2322851 127 + 2474545 ---------------GGUGGAGGACGCGUCCAGCUGUGC-CGCAGCGCCUCGUGUGUCAAUGGCCACACACGAACGCCAUUUGGCGC---UGGGUGUUGAUGGCCAGCUCAGAGAG-GGGGGCCAGGU-GCGGCAACAUAUCCACGAA------- ---------------.((((((((....))).((((..(-(.(((((((((((((((........)))))))).((((....)))).---..))))))).(((((..(((.....)-)).))))))).-.))))......)))))...------- ( -63.90, z-score = -2.92, R) >droMoj3.scaffold_6496 1681322 123 + 26866924 ------------GGUGGACGACGUCGCAUCCAGCUGUGC-AGCAGCGCCUCGUGUGUCAAUGGCCACACACGAGCCCCAUUUGGAGC---UGGGUGUUGAUGGCCAAUGGCGAAGUUAGAGUCGAUAC-AUAUACACAGA--------------- ------------.((.(((..((((((((((((((((((-....))))(((((((((........)))))))))..((....)))))---)))))).)))))(((...))).........))).))..-...........--------------- ( -46.10, z-score = -1.25, R) >droGri2.scaffold_15245 2570231 149 - 18325388 GGAGGGGGGCGUGAUUGGAGUCGGCACAUCCAGCUGUGC-GGCAGCGCCUCGUGUGUCAAUGGCCACACACGAGCGCCAUUUGGCGA---UGGGUGUUGAUGGUAUAUGCAAGAGUUGCUGAGAUGCC-GCAGAAAAAGUAGCCGA-GCUACAGU ((..(((.((.((((....))))))...)))..)).(((-((((((((.((((((((........))))))))))))..((..((((---(...(((...........)))...)))))..)).))))-)))......(((((...-)))))... ( -55.30, z-score = -0.34, R) >consensus ____________GGUUGAGGAGAACGCAUCCAGCUGUGC_UGCAGCGCCUCGUGUGUCACUUGCCACACACGAACGCCAUUUGGCGA___UUGGUGUUGAUGGCCAAUGCAGCGAGCAGAGACCAAAG_GCAGGAGGAGAUCCGGGCGCA_____ ................................((...((...((.((((((((((((........)))))))).((((....))))......)))).))...))....))............................................. (-24.52 = -24.74 + 0.22)


| Location | 12,185,114 – 12,185,251 |
|---|---|
| Length | 137 |
| Sequences | 12 |
| Columns | 155 |
| Reading direction | reverse |
| Mean pairwise identity | 70.52 |
| Shannon entropy | 0.61454 |
| G+C content | 0.58083 |
| Mean single sequence MFE | -44.77 |
| Consensus MFE | -23.55 |
| Energy contribution | -23.94 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.19 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.744830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
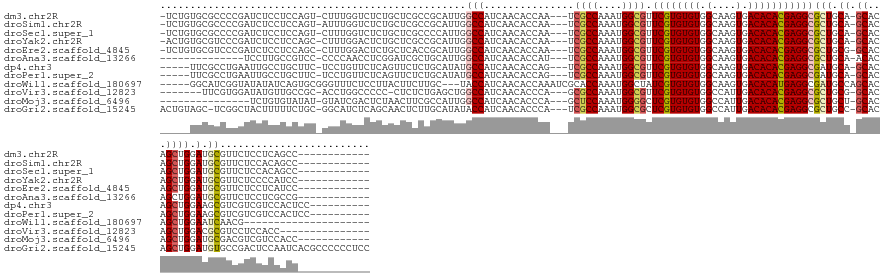
>dm3.chr2R 12185114 137 - 21146708 -UCUGUGCGCCCCGAUCUCCUCCAGU-CUUUGGUCUCUGCUCGCCGCAUUGGCCAUCAACACCAA---UCGCCAAAUGGCGUUCGUGUGUGGCAAGUGACACACGAGGCGCUGCA-GCACAGCUGGAUGCGUUCUCCUCAGCC------------ -.(((((((((.............((-...(((((..(((.....)))..)))))...)).....---.((((....)))).((((((((.(....).))))))))))))).)))-)....(((((...........))))).------------ ( -46.90, z-score = -0.52, R) >droSim1.chr2R 10917755 137 - 19596830 -UCUGUGCGCCCCGAUCUCCUCCAGU-AUUUGGUCUCUGCUCGCCGCAUUGGCCAUCAACACCAA---UCGCCAAAUGGCGUUCGUGUGUGGCAAGUGACACACGAGGCGCUGCA-GCACAGCUGGAUGCGUUCUCCACAGCC------------ -.(((((((((.............((-...(((((..(((.....)))..)))))...)).....---.((((....)))).((((((((.(....).))))))))))))).)))-)....((((.............)))).------------ ( -47.12, z-score = -0.48, R) >droSec1.super_1 9684486 137 - 14215200 -UCUGUGCGCCCCGAUCUCCUCCAGU-CUUUGGUCUCUGCUCGCCCCAUUGGCCAUCAACACCAA---UCGCCAAAUGGCGUUCGUGUGUGGCAAGUGACACACGAGGCGCUGCA-GCACAGCUGGAUGCGUUCUCCACAGCC------------ -.(((((.(....((.(.(.((((((-...((....((((.((((((((((((.((........)---).))).)))))...((((((((.(....).))))))))))))..)))-)..)))))))).).).)).))))))..------------ ( -45.60, z-score = -0.53, R) >droYak2.chr2R 4338155 137 + 21139217 -ACUGUGCGUCCCGAUCUCCUCCAGC-CUUUGGACUCUGCUCGCCGCAUUGGCCAUCAACACCAA---UCGCCAAAUGGCGUUCGUGUGUGGCAAGUGACACACGAGGCGCUGCA-GCACAGCUGGAUGCGUUCUCCCCAUCC------------ -.((((((((((.((...........-.)).))))..(((.((((..(((((.........))))---)((((....)))).((((((((.(....).))))))))))))..)))-))))))..(((((.(......))))))------------ ( -48.10, z-score = -1.17, R) >droEre2.scaffold_4845 8912379 137 + 22589142 -UCUGUGCGUCCCGAUCUCCUCCAGC-CUUUGGACUCUGCUCACCGCAUUGGCCAUCAACACCAA---UCGCCAAAUGGCGUUCGUGUGUGGCAAGUGACACACGAGGCGCUGCG-GCACAGCUGGAUGCGUUCUCCUCAUCC------------ -.....((((((........(((((.-..)))))....(((..(((((...(((...........---.((((....)))).((((((((.(....).)))))))))))..))))-)...))).)))))).............------------ ( -46.40, z-score = -0.77, R) >droAna3.scaffold_13266 15564081 124 - 19884421 --------------UCCUUGCCGUCC-CCCCAACCUCGGAUCGCUGCAUUGGCCAUCAACACCAU---UCGCCAAAUGGCGUUCGUGUGUGGCAAGUGACACACGAGGCGCUGCA-ACACAGCUGGAUGCGUUCUCCUCGCCG------------ --------------.....(((((((-......(((((..(((((......(((((..((((...---.((((....))))...))))))))).)))))....))))).((((..-...)))).))))).))...........------------ ( -41.00, z-score = -0.79, R) >dp4.chr3 15815486 135 + 19779522 -----UUCGCCUGAAUUGCCUGCUUC-UCCUGUUCUCAGUUCUCUGCAUAUGCCAUCAACACCAG---UCGCCAAAUGGCGUUCGUGUGUGGCAAGUGACACACGAGGCGAUGCA-GCACAGCUGGAAGCGUCGUCGUCCACUCC---------- -----...((..((((((...((...-....))...))))))...))...((((((..((((.((---.((((....)))))).))))))))))((((((..((((.((..(.((-((...)))).).)).)))).)).))))..---------- ( -43.50, z-score = -0.75, R) >droPer1.super_2 4888340 135 + 9036312 -----UUCGCCUGAAUUGCCUGCUUC-UCCUGUUCUCAGUUCUCUGCAUAUGCCAUCAACACCAG---UCGCCAAAUGGCGUUCGUGUGUGGCAAGUGACACACGAGGCGAUGCA-GCACAGCUGGAAGCGUCGUCGUCCACUCC---------- -----...((..((((((...((...-....))...))))))...))...((((((..((((.((---.((((....)))))).))))))))))((((((..((((.((..(.((-((...)))).).)).)))).)).))))..---------- ( -43.50, z-score = -0.75, R) >droWil1.scaffold_180697 1752621 126 - 4168966 -----GGCAUCGGUAUAUAUCAGUGCGGGUUUCUCCUUACUUCUUGC---UACCAUCAACACCAAAUCGCACCAAAUGGCUAUCGUGUGUGGCAAGUGACACAUGAGGCGAUGCCAGCACAGCUGGAAUCAACG--------------------- -----(((...(((....))).((((.(((.((.(((((...(((((---((((((.....(((............))).....))).)))))))).......))))).)).))).)))).)))..........--------------------- ( -34.30, z-score = -0.16, R) >droVir3.scaffold_12823 2322851 127 - 2474545 -------UUCGUGGAUAUGUUGCCGC-ACCUGGCCCCC-CUCUCUGAGCUGGCCAUCAACACCCA---GCGCCAAAUGGCGUUCGUGUGUGGCCAUUGACACACGAGGCGCUGCG-GCACAGCUGGACGCGUCCUCCACC--------------- -------...(((((..(((.(((((-(.((((((...-(((...)))..))))).......((.---(((((....)))))((((((((........)))))))))).).))))-)))))...(((....)))))))).--------------- ( -52.10, z-score = -1.71, R) >droMoj3.scaffold_6496 1681322 123 - 26866924 ---------------UCUGUGUAUAU-GUAUCGACUCUAACUUCGCCAUUGGCCAUCAACACCCA---GCUCCAAAUGGGGCUCGUGUGUGGCCAUUGACACACGAGGCGCUGCU-GCACAGCUGGAUGCGACGUCGUCCACC------------ ---------------.(((((((...-(((.((.(((.....(((....(((((((..((((..(---(((((....)))))).))))))))))).))).....))).)).))))-)))))).((((((......))))))..------------ ( -43.70, z-score = -1.50, R) >droGri2.scaffold_15245 2570231 149 + 18325388 ACUGUAGC-UCGGCUACUUUUUCUGC-GGCAUCUCAGCAACUCUUGCAUAUACCAUCAACACCCA---UCGCCAAAUGGCGCUCGUGUGUGGCCAUUGACACACGAGGCGCUGCC-GCACAGCUGGAUGUGCCGACUCCAAUCACGCCCCCCUCC ..((.((.-(((((((((..((.(((-((((.(.(.(((.....)))..................---.((((....))))(((((((((........)))))))))).).))))-))).))..)).)).))))))).))............... ( -45.00, z-score = -1.02, R) >consensus _____UGCGCCCCGAUCUCCUCCUGC_CUUUGGUCUCUGCUCGCUGCAUUGGCCAUCAACACCAA___UCGCCAAAUGGCGUUCGUGUGUGGCAAGUGACACACGAGGCGCUGCA_GCACAGCUGGAUGCGUCCUCCUCAACC____________ .............................................(((...(((...............((((....)))).((((((((.(....).)))))))))))..))).......((.....))......................... (-23.55 = -23.94 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:49 2011