| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,129,719 – 12,129,826 |
| Length | 107 |
| Max. P | 0.919163 |

| Location | 12,129,719 – 12,129,826 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.41 |
| Shannon entropy | 0.37286 |
| G+C content | 0.48297 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -17.76 |
| Energy contribution | -17.53 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12129719 107 - 21146708 ----------AUGA-AGGGGUCGGCGGGGGGUGUCGAAAAAUGACUGACAAUAAAAUGUUGACGCAGCCAAGGAGCAUUAUGG--AUACAGGGUGCUCGAAGGCGAAUGGAUUAAGUCGA ----------.(((-..(.(((((((.....(((((.........)))))......))))))).).(((...((((((..((.--...))..))))))...)))............))). ( -30.40, z-score = -2.72, R) >droSim1.chr2R 10863016 107 - 19596830 ----------AUGA-AGGGGACGGCGGGGGGUGUCGGAAAAUGACUGACAAUAAAAUGUUGACGCAGCCAAAGAGCAUUAUGG--AUACAGGGUGCUCGAAGGCGAUUGGAUUAAGUCGA ----------....-..(.(.(((((.....((((((.......))))))......))))).).).(((...((((((..((.--...))..))))))...)))((((......)))).. ( -31.40, z-score = -3.33, R) >droSec1.super_1 9629188 107 - 14215200 ----------AUGA-AGGGGACGGCGGGGGGUGUCGGAAAAUGACUGACAAUAAAAUGUUGACGCAGCCAAAGAGCAUUAUGG--AUACAGGGUGCUCGAAGGCGAUUUGAUUAAGUCGA ----------....-..(.(.(((((.....((((((.......))))))......))))).).).(((...((((((..((.--...))..))))))...)))(((((....))))).. ( -30.90, z-score = -3.02, R) >droYak2.chr2R 4286795 118 + 21139217 GAGGGACGGUGUGCGGGGGGUGUGCGGGGAGUGUCGGAAAAUGACUGACAAUAAAAUGUUGACGCAGCCAAAGAGCAUUAUGA--AUACAGGGUGCUCGAAGGCGAUUGGAUUAAGUCGA ....(((...((.(((...(((((((.....((((((.......))))))......)))..)))).(((...((((((..((.--...))..))))))...)))..))).))...))).. ( -32.90, z-score = -2.06, R) >droEre2.scaffold_4845 8861001 108 + 22589142 ----------GUGAGGGGGGCUGGCGGGGGGUGUCGGAAAAUGACUGACAAUAAAAUGUUGACGCAGCCAACGAGCAUUAUGG--AUACAGGGUGCUCGAAGGCGAUUGGAUUAAGUCGA ----------........(((((((((.(..((((((.......))))))......).))).).)))))..(((((((..((.--...))..)))))))....(((((......))))). ( -33.80, z-score = -2.99, R) >droAna3.scaffold_13266 15507021 95 - 19884421 -----------------------CUGGGGUGUGCUGGAAAAUGACUGACAAUAAAAUGUUGACGCAGCCAAAGAGCAUUAUGG--AUACAGGGUGCUCGAAGGCGAUUGGAUUAAGUCGA -----------------------(..(......)..)....(((((..((((....(((....)))(((...((((((..((.--...))..))))))...))).)))).....))))). ( -25.10, z-score = -1.76, R) >droPer1.super_2 4833666 105 + 9036312 ---------------ACGACGGGGUGCCUGCCGGUAGAAAAUGACUGACAAUAAAAUGUUGACGCAGUCAAAGAGCAUUAUGAAAAUACAGGGUGACUCAAAGCGAUUGGAUUAAGUGGA ---------------.(.((..(((.((...((.(......((((((.((((.....))))...))))))..((((((..((......))..))).)))..).))...)))))..)).). ( -22.60, z-score = -0.50, R) >dp4.chr3 15762851 102 + 19779522 ---------------ACGACGGGGUGCCUGCCGGUAGAAAAUGACUGACAAUAAAAUGUUGACGCAGUCAAAGAGCAUUAUGAAUAUACAGGGGGACUCAAAGCGAUUGGAUUAAGU--- ---------------.((...((((.(((.((.(((.....((((((.((((.....))))...)))))).....(.....)....))).)))))))))....))............--- ( -20.10, z-score = 0.00, R) >consensus ___________UGA_AGGGGACGGCGGGGGGUGUCGGAAAAUGACUGACAAUAAAAUGUUGACGCAGCCAAAGAGCAUUAUGG__AUACAGGGUGCUCGAAGGCGAUUGGAUUAAGUCGA ........................(((......))).....(((((((((......))))......(((...((((((..((......))..))))))...)))..........))))). (-17.76 = -17.53 + -0.23)

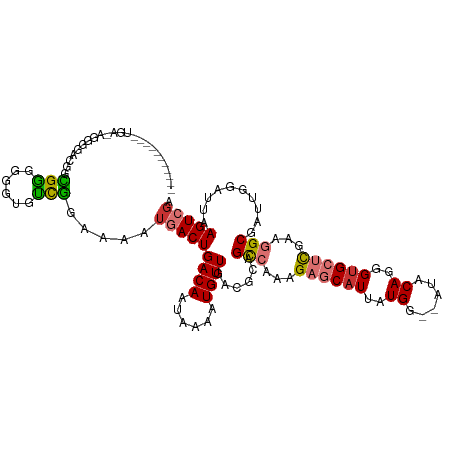
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:47 2011