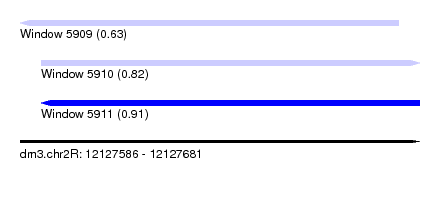
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,127,586 – 12,127,681 |
| Length | 95 |
| Max. P | 0.912803 |

| Location | 12,127,586 – 12,127,676 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 77.78 |
| Shannon entropy | 0.30610 |
| G+C content | 0.45116 |
| Mean single sequence MFE | -26.37 |
| Consensus MFE | -15.76 |
| Energy contribution | -17.10 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.627160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12127586 90 - 21146708 GUCAAUGAAAACCUCUCAGAAGAG---AGAGACGCUACUCCGCAGCCUUUAGCAGUUUCCAAGAACAUGGAG---GAGACUAUGCCCUCGAAUCCU (((..(....).(((((....)))---)).)))....((((((........))....((((......)))))---))).................. ( -18.70, z-score = 0.18, R) >droSim1.chr2R_random 2505451 96 - 2996586 GUCAAUGAAAACCUCUCAAAACAGUGUAAGGCUGCUACUCUGCAGCCUUUGGAAAGUUCCAAGAACAUGGAACUACAAGUAGAGGAGACUAUGCCU (((..(....)((((((..........((((((((......)))))))).....(((((((......)))))))....).))))).)))....... ( -30.20, z-score = -2.92, R) >droSec1.super_1 9627071 96 - 14215200 GUCAAUGAAAACCUCUCAAAAGAGUGUAAGGCUGCUACUCUGCAGCCUUUGGAAAGUUCCAAGAACAUGGAACUACAAGUAAAGGAGACUAUGCCU ..........((((((....)))).))((((((((......)))))))).....(((((((......)))))))....(((.((....)).))).. ( -30.20, z-score = -3.19, R) >consensus GUCAAUGAAAACCUCUCAAAAGAGUGUAAGGCUGCUACUCUGCAGCCUUUGGAAAGUUCCAAGAACAUGGAACUACAAGUAAAGGAGACUAUGCCU ............((((....))))...((((((((......)))))))).....(((((((......)))))))....(((.((....)).))).. (-15.76 = -17.10 + 1.34)

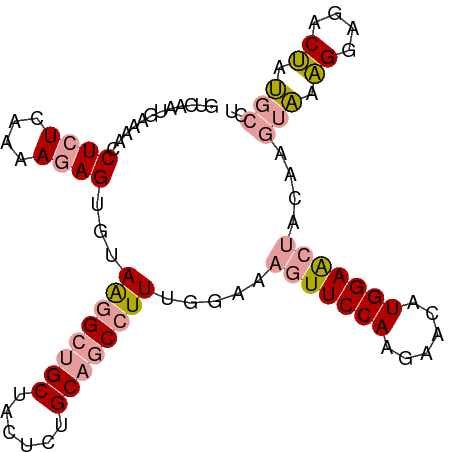
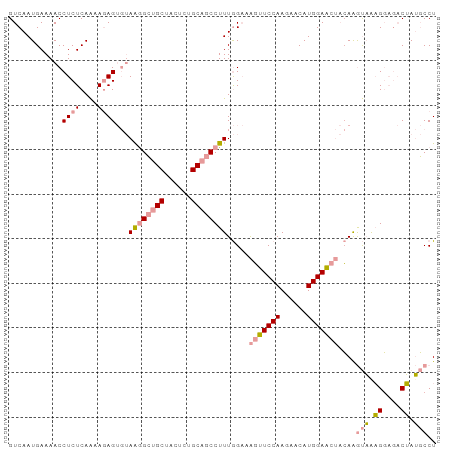
| Location | 12,127,591 – 12,127,681 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.03 |
| Shannon entropy | 0.28621 |
| G+C content | 0.46000 |
| Mean single sequence MFE | -29.61 |
| Consensus MFE | -21.79 |
| Energy contribution | -22.80 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.819967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12127591 90 + 21146708 UCGAGGGCAUAGUCUCCU------------CCAUGUUCUUGGAAACUGCUAAAGGCUGCGGAGUAGCGUCUCU---CUCUUCUGAGAGGUUUUCAUUGACCACUG ..(((((.......))))------------)...(((..(((((((........(((((...)))))..((((---(......))))))))))))..)))..... ( -25.10, z-score = -0.13, R) >droSim1.chr2R_random 2505452 100 + 2996586 -----GGCAUAGUCUCCUCUACUUGUAGUUCCAUGUUCUUGGAACUUUCCAAAGGCUGCAGAGUAGCAGCCUUACACUGUUUUGAGAGGUUUUCAUUGACCACUG -----((((......(((((...((.(((((((......)))))))...))((((((((......))))))))...........))))).......)).)).... ( -32.52, z-score = -2.65, R) >droSec1.super_1 9627072 100 + 14215200 -----GGCAUAGUCUCCUUUACUUGUAGUUCCAUGUUCUUGGAACUUUCCAAAGGCUGCAGAGUAGCAGCCUUACACUCUUUUGAGAGGUUUUCAUUGACUACUG -----....(((((......(((((((((((((......))))))).....((((((((......)))))))))).(((....))))))).......)))))... ( -31.22, z-score = -2.68, R) >consensus _____GGCAUAGUCUCCU_UACUUGUAGUUCCAUGUUCUUGGAACUUUCCAAAGGCUGCAGAGUAGCAGCCUUACACUCUUUUGAGAGGUUUUCAUUGACCACUG ............((((...........((((((......))))))......((((((((......))))))))..........))))((((......)))).... (-21.79 = -22.80 + 1.01)

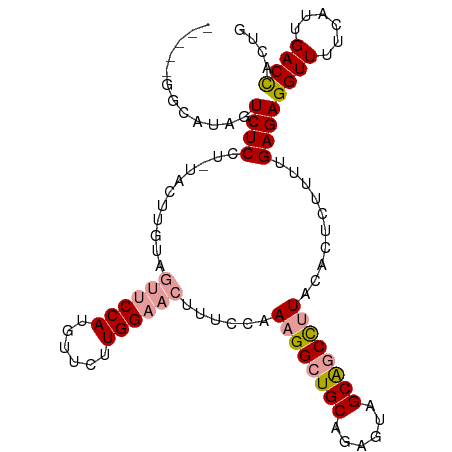
| Location | 12,127,591 – 12,127,681 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.03 |
| Shannon entropy | 0.28621 |
| G+C content | 0.46000 |
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -19.64 |
| Energy contribution | -21.53 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.912803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
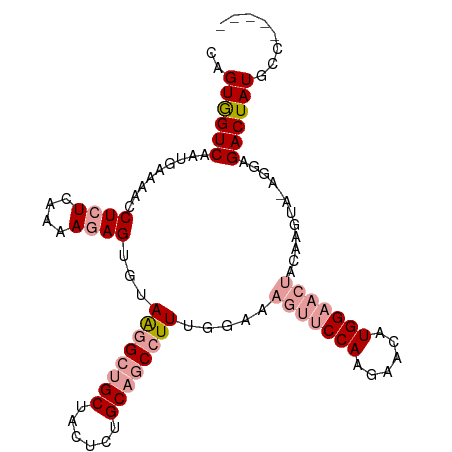
>dm3.chr2R 12127591 90 - 21146708 CAGUGGUCAAUGAAAACCUCUCAGAAGAG---AGAGACGCUACUCCGCAGCCUUUAGCAGUUUCCAAGAACAUGG------------AGGAGACUAUGCCCUCGA ..((((((.........(((((......)---))))......((((((........)).((((....))))..))------------))..))))))........ ( -22.30, z-score = -0.20, R) >droSim1.chr2R_random 2505452 100 - 2996586 CAGUGGUCAAUGAAAACCUCUCAAAACAGUGUAAGGCUGCUACUCUGCAGCCUUUGGAAAGUUCCAAGAACAUGGAACUACAAGUAGAGGAGACUAUGCC----- ..((((((..(....)((((((..........((((((((......)))))))).....(((((((......)))))))....).))))).))))))...----- ( -34.00, z-score = -3.27, R) >droSec1.super_1 9627072 100 - 14215200 CAGUAGUCAAUGAAAACCUCUCAAAAGAGUGUAAGGCUGCUACUCUGCAGCCUUUGGAAAGUUCCAAGAACAUGGAACUACAAGUAAAGGAGACUAUGCC----- ..((((((.......((((((....)))).))((((((((......)))))))).....(((((((......)))))))............))))))...----- ( -32.30, z-score = -3.47, R) >consensus CAGUGGUCAAUGAAAACCUCUCAAAAGAGUGUAAGGCUGCUACUCUGCAGCCUUUGGAAAGUUCCAAGAACAUGGAACUACAAGUA_AGGAGACUAUGCC_____ ..((((((........................((((((((......)))))))).....(((((((......)))))))............))))))........ (-19.64 = -21.53 + 1.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:45 2011