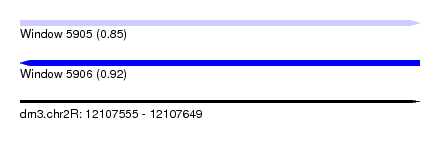
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,107,555 – 12,107,649 |
| Length | 94 |
| Max. P | 0.921796 |

| Location | 12,107,555 – 12,107,649 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.72 |
| Shannon entropy | 0.38001 |
| G+C content | 0.47140 |
| Mean single sequence MFE | -29.94 |
| Consensus MFE | -28.38 |
| Energy contribution | -28.50 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.04 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12107555 94 + 21146708 CUCCCAUAUGGUCUAGUGGCUAGGAUAUCUGGCUUUCACCCAGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAGAGUACUUUUUUUUUACUGCU-------------- .((((((((((((....)))).........(((((((.....)))))))((((.......))))))))))))......................-------------- ( -29.40, z-score = -0.65, R) >droSim1.chr2R 10847163 94 + 19596830 CUCCCAUAUGGUCUAGUGGCUAGGAUAUCUGGCUUUCACCCAGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAGAGUGCUUUUUUUUUACUGCU-------------- .((((((((((((....)))).........(((((((.....)))))))((((.......))))))))))))......................-------------- ( -29.40, z-score = -0.45, R) >droSec1.super_1 9606605 104 + 14215200 CUCCCAUAUGGUCUAGUGGCUAGGAUAUCUGGCUUUCACCCAGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAGAGUACUUUUUUUUUAUAUUUGUUUACUGCU---- .((((((((((((....)))).........(((((((.....)))))))((((.......))))))))))))................................---- ( -29.40, z-score = -0.47, R) >droEre2.scaffold_4845 8838271 102 - 22589142 CUCCCAUAUGGUCUAGUGGCUAGGAUAUCUGGCUUUCACCCAGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAG--UACAUUUUUUUAAUAUUUUUUUGCUGCC---- .((((((((((((....)))).........(((((((.....)))))))((((.......))))))))))))..--............................---- ( -29.40, z-score = -0.71, R) >dp4.chr3 4470450 100 + 19779522 AUCCCAUAUGGUCUAGUGGCUAGGAUAUCUGGCUUUCACCCAGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAGAACACUUUUUUCAUUCUCUUUUUAUU-------- .((((((((((((....)))).........(((((((.....)))))))((((.......))))))))))))............................-------- ( -29.60, z-score = -1.13, R) >droPer1.super_2 4658847 100 + 9036312 AUCCCAUAUGGUCUAGUGGCUAGGAUAUCUGGCUUUCACCCAGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAGAACACUUUUUUUAUUCUCUUUUUAUU-------- .((((((((((((....)))).........(((((((.....)))))))((((.......))))))))))))............................-------- ( -29.60, z-score = -1.19, R) >droWil1.scaffold_180700 4041821 90 + 6630534 AUCCCAUAUGGUCUAGUGGCUAGGAUAUCUGGCUUUCACCCAGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAAU-UAUUUUUUUCCAACU----------------- .((((((((((((....)))).........(((((((.....)))))))((((.......))))))))))))...-...............----------------- ( -29.60, z-score = -1.44, R) >droVir3.scaffold_12875 15833148 108 - 20611582 CUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAGCAUACCAAACUGAGUAGAUCGGGUAAUUUUUUGG ...((((........))))......((((((((((((((((((.....)))))).........((((((.....)))))).....))).)).)))))))......... ( -33.70, z-score = -0.49, R) >anoGam1.chr2L 2157521 93 - 48795086 CUCCCAUAUGGUCUAGUGGCUAGGAUAUCUGGCUUUCACCCAGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAACA-UACCUUUUUUCUUUUGUU-------------- .((((((((((((....)))).........(((((((.....)))))))((((.......))))))))))))...-..................-------------- ( -29.40, z-score = -1.14, R) >consensus CUCCCAUAUGGUCUAGUGGCUAGGAUAUCUGGCUUUCACCCAGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAGA_UACUUUUUUUUUACUGUU______________ .((((((((((((....)))).........(((((((.....)))))))((((.......)))))))))))).................................... (-28.38 = -28.50 + 0.12)

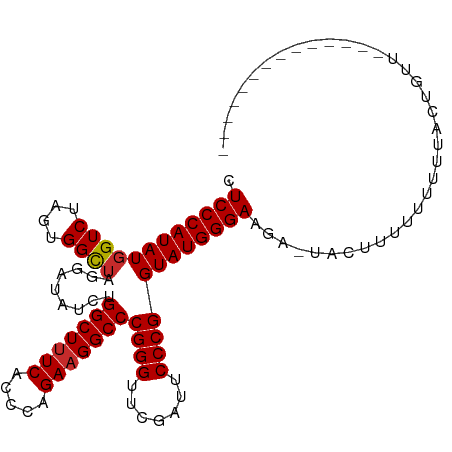
| Location | 12,107,555 – 12,107,649 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 81.72 |
| Shannon entropy | 0.38001 |
| G+C content | 0.47140 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -25.85 |
| Energy contribution | -25.77 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.921796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12107555 94 - 21146708 --------------AGCAGUAAAAAAAAAGUACUCUUCCCAUACCGGGAAUCGAACCCGGGCCUUCUGGGUGAAAGCCAGAUAUCCUAGCCACUAGACCAUAUGGGAG --------------......................((((((((((((.......)))))....(((((.(....))))))....((((...))))....))))))). ( -25.90, z-score = -1.03, R) >droSim1.chr2R 10847163 94 - 19596830 --------------AGCAGUAAAAAAAAAGCACUCUUCCCAUACCGGGAAUCGAACCCGGGCCUUCUGGGUGAAAGCCAGAUAUCCUAGCCACUAGACCAUAUGGGAG --------------.((............)).....((((((((((((.......)))))....(((((.(....))))))....((((...))))....))))))). ( -26.00, z-score = -1.03, R) >droSec1.super_1 9606605 104 - 14215200 ----AGCAGUAAACAAAUAUAAAAAAAAAGUACUCUUCCCAUACCGGGAAUCGAACCCGGGCCUUCUGGGUGAAAGCCAGAUAUCCUAGCCACUAGACCAUAUGGGAG ----................................((((((((((((.......)))))....(((((.(....))))))....((((...))))....))))))). ( -25.90, z-score = -1.13, R) >droEre2.scaffold_4845 8838271 102 + 22589142 ----GGCAGCAAAAAAAUAUUAAAAAAAUGUA--CUUCCCAUACCGGGAAUCGAACCCGGGCCUUCUGGGUGAAAGCCAGAUAUCCUAGCCACUAGACCAUAUGGGAG ----............................--..((((((((((((.......)))))....(((((.(....))))))....((((...))))....))))))). ( -25.90, z-score = -0.85, R) >dp4.chr3 4470450 100 - 19779522 --------AAUAAAAAGAGAAUGAAAAAAGUGUUCUUCCCAUACCGGGAAUCGAACCCGGGCCUUCUGGGUGAAAGCCAGAUAUCCUAGCCACUAGACCAUAUGGGAU --------.........(((((.........)))))((((((((((((.......)))))....(((((.(....))))))....((((...))))....))))))). ( -26.80, z-score = -1.17, R) >droPer1.super_2 4658847 100 - 9036312 --------AAUAAAAAGAGAAUAAAAAAAGUGUUCUUCCCAUACCGGGAAUCGAACCCGGGCCUUCUGGGUGAAAGCCAGAUAUCCUAGCCACUAGACCAUAUGGGAU --------.........((((((.......))))))((((((((((((.......)))))....(((((.(....))))))....((((...))))....))))))). ( -27.90, z-score = -1.71, R) >droWil1.scaffold_180700 4041821 90 - 6630534 -----------------AGUUGGAAAAAAAUA-AUUUCCCAUACCGGGAAUCGAACCCGGGCCUUCUGGGUGAAAGCCAGAUAUCCUAGCCACUAGACCAUAUGGGAU -----------------...............-...((((((((((((.......)))))....(((((.(....))))))....((((...))))....))))))). ( -25.70, z-score = -0.87, R) >droVir3.scaffold_12875 15833148 108 + 20611582 CCAAAAAAUUACCCGAUCUACUCAGUUUGGUAUGCUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAG .........((((.(((.......))).))))....(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....))))))). ( -31.40, z-score = -0.98, R) >anoGam1.chr2L 2157521 93 + 48795086 --------------AACAAAAGAAAAAAGGUA-UGUUCCCAUACCGGGAAUCGAACCCGGGCCUUCUGGGUGAAAGCCAGAUAUCCUAGCCACUAGACCAUAUGGGAG --------------..................-...((((((((((((.......)))))....(((((.(....))))))....((((...))))....))))))). ( -25.90, z-score = -0.96, R) >consensus ______________AACAGUAAAAAAAAAGUA_UCUUCCCAUACCGGGAAUCGAACCCGGGCCUUCUGGGUGAAAGCCAGAUAUCCUAGCCACUAGACCAUAUGGGAG ....................................((((((((((((.......)))))....(((((.(....))))))....((((...))))....))))))). (-25.85 = -25.77 + -0.09)
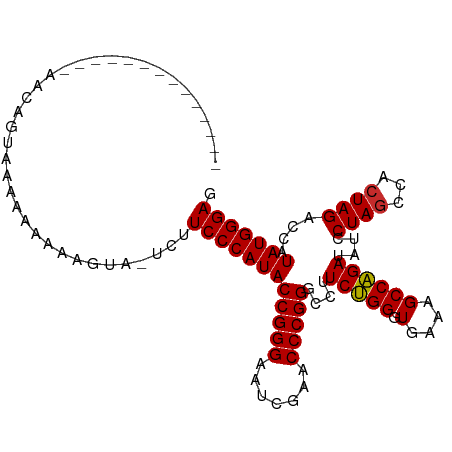
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:41 2011