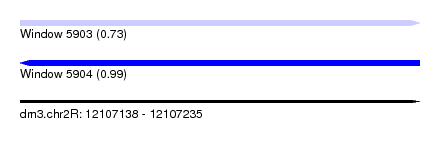
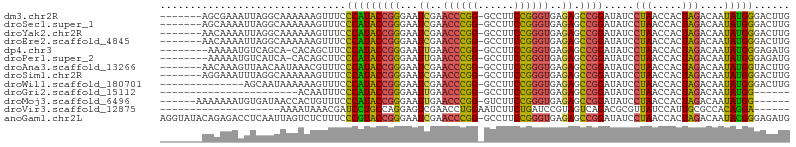
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,107,138 – 12,107,235 |
| Length | 97 |
| Max. P | 0.989896 |

| Location | 12,107,138 – 12,107,235 |
|---|---|
| Length | 97 |
| Sequences | 13 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.32 |
| Shannon entropy | 0.50606 |
| G+C content | 0.49762 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -22.89 |
| Energy contribution | -21.72 |
| Covariance contribution | -1.18 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.52 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.728568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
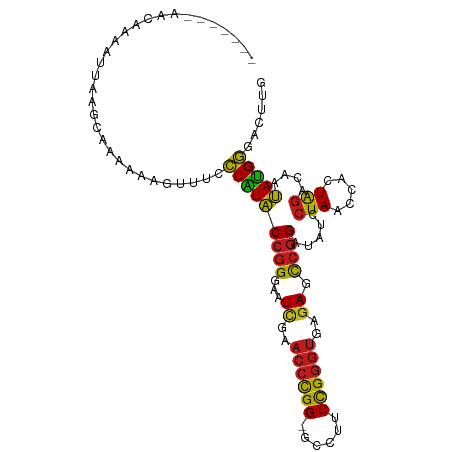
>dm3.chr2R 12107138 97 + 21146708 CAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGC-CCGGGUUCGAUUCCCGGUAUGGGAAACUUUUUUGCCUAAUUUCGCU------- ....(((((((.(((((........))))).((((.((..((((((.....-))))))..))...)))))))))))......................------- ( -28.80, z-score = -0.35, R) >droSec1.super_1 9606280 97 + 14215200 CAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGC-CCGGGUUCGAUUCCCGGUAUGGGAAACUUUUUUGCCUAAUUUUGCU------- ....(((((((.(((((........))))).((((.((..((((((.....-))))))..))...)))))))))))......................------- ( -28.80, z-score = -0.39, R) >droYak2.chr2R 4263609 97 - 21139217 CAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGC-CCGGGUUCGAUUCCCGGUAUGGGAAACUUUUUUGCCUAAUUUUGUU------- ....(((((((.(((((........))))).((((.((..((((((.....-))))))..))...)))))))))))......................------- ( -28.80, z-score = -0.57, R) >droEre2.scaffold_4845 8837960 97 - 22589142 CAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGC-CCGGGUUCGAUUCCCGGUAUGGGAAACUUUUUUGCCUAAUUUUGUU------- ....(((((((.(((((........))))).((((.((..((((((.....-))))))..))...)))))))))))......................------- ( -28.80, z-score = -0.57, R) >dp4.chr3 7003016 95 - 19779522 CAUCUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGC-CCGGGUUCAAUUCCCGGUAUGGGAAGCUGUG-UGCUGACAUUUUU-------- ....(((((((.(((((........))))).((((.....((((((.....-)))))).......)))))))))))(((....-.))).........-------- ( -29.80, z-score = -0.51, R) >droPer1.super_2 8848818 95 + 9036312 CAUCUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGC-CCGGGUUCAAUUCCCGGUAUGGGAAGCUGUG-UGAUGACAUUUUU-------- (((((((((((.(((((........))))).((((.....((((((.....-)))))).......))))))))))).......-.))))........-------- ( -28.50, z-score = -0.13, R) >droAna3.scaffold_13266 15715813 97 - 19884421 CAAGUACCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGC-CCGGGUUCGAUUCCCGGUAUGGGAAACGUUUAUUGUUAACUUUGUU------- .(((((((((........)))))...((((..((..(((((.((....)).-(((((.......)))))..)))))..))..))))....))))....------- ( -27.80, z-score = -0.46, R) >droSim1.chr2R 10846833 97 + 19596830 CAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGC-CCGGGUUCGAUUCCCGGUAUGGGAAACUUUUUUGCCUAAAUUUCCU------- ....(((((((.(((((........))))).((((.((..((((((.....-))))))..))...)))))))))))......................------- ( -28.80, z-score = -0.45, R) >droWil1.scaffold_180701 2125502 90 - 3904529 CAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGC-CCGGGUUCGAUUCCCGGUAUGGGAAACUUUUUUAUUGCU-------------- ....(((((((.(((((........))))).((((.((..((((((.....-))))))..))...)))))))))))...............-------------- ( -28.80, z-score = -0.93, R) >droGri2.scaffold_15112 2182152 75 + 5172618 ------CCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGC-CCGGGUUCAAUUCCCGGUAUGGGAAAUUGU----------------------- ------(((((.(((((........))))).((((.....((((((.....-)))))).......)))))))))........----------------------- ( -22.60, z-score = 0.03, R) >droMoj3.scaffold_6496 5841950 92 - 26866924 ------CCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGAC-CCGGGUUCAAUUCCCGGUAUGGGAAACAGUGGUUAUCACAUUUUUUU------ ------((((........))))....((((.(((.((...((((((.....-))))))....(((((.....)))))..))))).))))..........------ ( -24.90, z-score = -0.16, R) >droVir3.scaffold_12875 15822436 79 - 20611582 ------UCCUGUGGCGCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGUUUAUUUU-------------------- ------(((((((((((..........))))).......(((((....)))))((((.......))))))))))...........-------------------- ( -22.10, z-score = -0.59, R) >anoGam1.chr2L 1818862 104 - 48795086 CAUCUCCCGUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGC-CCGGGUUCGAUUCCCGGUACGGGAAAGAGACUAAUUGAGGUCUCUGUAUACCU ....(((((((.(((((........))))).((((.((..((((((.....-))))))..))...))))))))))).(((((((......)))))))........ ( -39.00, z-score = -1.62, R) >consensus CAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGC_CCGGGUUCGAUUCCCGGUAUGGGAAACUUUUUUGCUUAAAUUUGUU_______ ......(((((.(((((........)))))(((((.......))))).....(((((.......))))))))))............................... (-22.89 = -21.72 + -1.18)
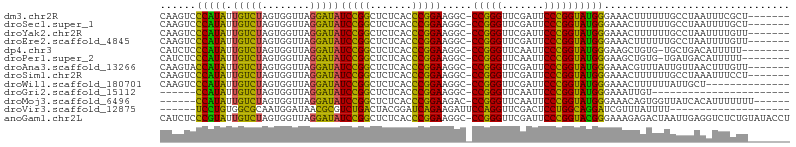
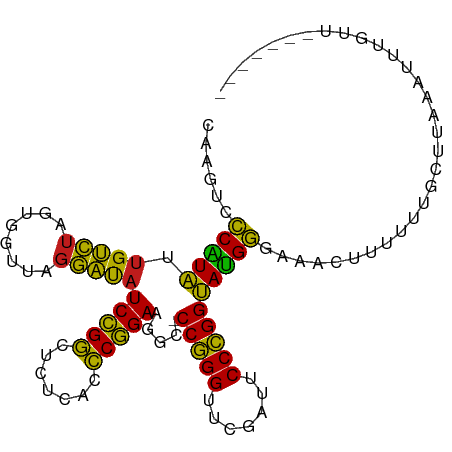
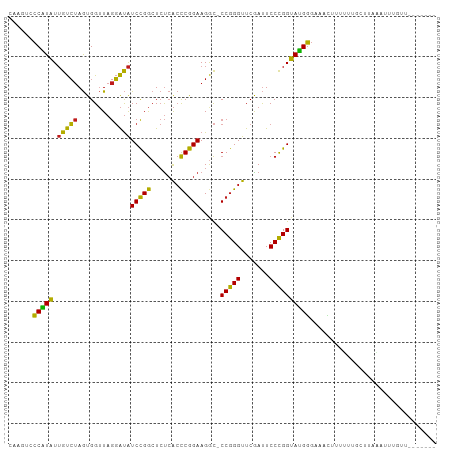
| Location | 12,107,138 – 12,107,235 |
|---|---|
| Length | 97 |
| Sequences | 13 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 77.32 |
| Shannon entropy | 0.50606 |
| G+C content | 0.49762 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -22.06 |
| Energy contribution | -21.57 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.989896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12107138 97 - 21146708 -------AGCGAAAUUAGGCAAAAAAGUUUCCCAUACCGGGAAUCGAACCCGG-GCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUG -------.((........)).........(((((((((((...((..((((((-.....))))))..)).)))).....(((.....)))....))))))).... ( -31.20, z-score = -1.79, R) >droSec1.super_1 9606280 97 - 14215200 -------AGCAAAAUUAGGCAAAAAAGUUUCCCAUACCGGGAAUCGAACCCGG-GCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUG -------.((........)).........(((((((((((...((..((((((-.....))))))..)).)))).....(((.....)))....))))))).... ( -31.00, z-score = -1.91, R) >droYak2.chr2R 4263609 97 + 21139217 -------AACAAAAUUAGGCAAAAAAGUUUCCCAUACCGGGAAUCGAACCCGG-GCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUG -------......................(((((((((((...((..((((((-.....))))))..)).)))).....(((.....)))....))))))).... ( -29.90, z-score = -1.86, R) >droEre2.scaffold_4845 8837960 97 + 22589142 -------AACAAAAUUAGGCAAAAAAGUUUCCCAUACCGGGAAUCGAACCCGG-GCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUG -------......................(((((((((((...((..((((((-.....))))))..)).)))).....(((.....)))....))))))).... ( -29.90, z-score = -1.86, R) >dp4.chr3 7003016 95 + 19779522 --------AAAAAUGUCAGCA-CACAGCUUCCCAUACCGGGAAUUGAACCCGG-GCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAGAUG --------......((((((.-....)))(((((((((((...((..((((((-.....))))))..)).)))).....(((.....)))....)))))))))). ( -31.20, z-score = -2.20, R) >droPer1.super_2 8848818 95 - 9036312 --------AAAAAUGUCAUCA-CACAGCUUCCCAUACCGGGAAUUGAACCCGG-GCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAGAUG --------.............-..((..((((((((((((...((..((((((-.....))))))..)).)))).....(((.....)))....)))))))).)) ( -29.00, z-score = -1.53, R) >droAna3.scaffold_13266 15715813 97 + 19884421 -------AACAAAGUUAACAAUAAACGUUUCCCAUACCGGGAAUCGAACCCGG-GCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGUACUUG -------..................((.(((((.....))))).)).((((((-.....))))))(((.((((......(((.....)))......)))).))). ( -25.30, z-score = -1.05, R) >droSim1.chr2R 10846833 97 - 19596830 -------AGGAAAUUUAGGCAAAAAAGUUUCCCAUACCGGGAAUCGAACCCGG-GCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUG -------((((.((((.(((........(((((.....)))))((..((((((-.....))))))..))))))))).))))..(((.((.....)))))...... ( -31.30, z-score = -1.57, R) >droWil1.scaffold_180701 2125502 90 + 3904529 --------------AGCAAUAAAAAAGUUUCCCAUACCGGGAAUCGAACCCGG-GCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUG --------------...............(((((((((((...((..((((((-.....))))))..)).)))).....(((.....)))....))))))).... ( -29.90, z-score = -2.32, R) >droGri2.scaffold_15112 2182152 75 - 5172618 -----------------------ACAAUUUCCCAUACCGGGAAUUGAACCCGG-GCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGG------ -----------------------........(((((((((...((..((((((-.....))))))..)).)))).....(((.....)))....)))))------ ( -22.50, z-score = -1.23, R) >droMoj3.scaffold_6496 5841950 92 + 26866924 ------AAAAAAAUGUGAUAACCACUGUUUCCCAUACCGGGAAUUGAACCCGG-GUCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGG------ ------........(((.....)))......((((((((((.......)))))-((((....(((....)))((....))........))))..)))))------ ( -24.60, z-score = -0.85, R) >droVir3.scaffold_12875 15822436 79 + 20611582 --------------------AAAAUAAACGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGA------ --------------------...........((((((((((.......))))).....((((((.....)))))).((((........))))..)))))------ ( -22.70, z-score = -1.94, R) >anoGam1.chr2L 1818862 104 + 48795086 AGGUAUACAGAGACCUCAAUUAGUCUCUUUCCCGUACCGGGAAUCGAACCCGG-GCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUACGGGAGAUG .........(((((........)))))(((((((((((((...((..((((((-.....))))))..)).)))).....(((.....)))....))))))))).. ( -39.10, z-score = -2.56, R) >consensus _______AACAAAAUUAAGCAAAAAAGUUUCCCAUACCGGGAAUCGAACCCGG_GCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUG ...............................(((((((((...((..((((((......))))))..)).)))).....(((.....)))....)))))...... (-22.06 = -21.57 + -0.49)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:40 2011