| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,095,138 – 12,095,232 |
| Length | 94 |
| Max. P | 0.989557 |

| Location | 12,095,138 – 12,095,232 |
|---|---|
| Length | 94 |
| Sequences | 13 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 85.31 |
| Shannon entropy | 0.33951 |
| G+C content | 0.51506 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -26.22 |
| Energy contribution | -25.72 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.989557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
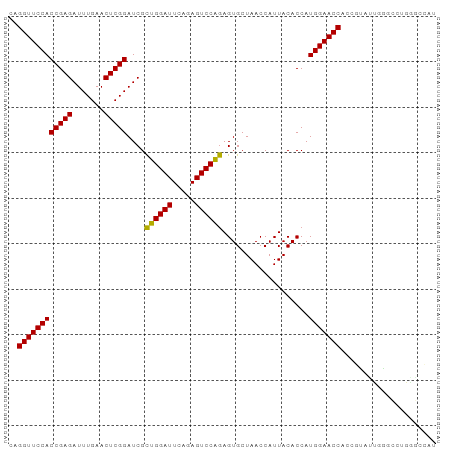
>dm3.chr2R 12095138 94 + 21146708 CAGGUUCCACCGAGAUUUGAACUCGGAUCGCUGGAUUCAGAGUCCAGAGUGCUAACCAUUACACCAUGGAACCACCGUAUUGGUCCUGGGCCAU ..((((((((((((.......)))))....((((((.....))))))...................))))))).......((((.....)))). ( -30.70, z-score = -1.68, R) >droSim1.chr2R 10834382 94 + 19596830 CAGGUUCCACCGAGAUUUGAACUCGGAUCGCUGGAUUCAGAGUCCAGAGUGCUAACCAUUACACCAUGGAACCACCGUAUUGGUCCUGGGCCAU ..((((((((((((.......)))))....((((((.....))))))...................))))))).......((((.....)))). ( -30.70, z-score = -1.68, R) >droSec1.super_1 9594335 94 + 14215200 CAGGUUCCACCGAGAUUUGAACUCGGAUCGCUGGAUUCAGAGUCCAGAGUGCUAACCAUUACACCAUGGAACCACCGUAUUGGUCCUGGGCCUU ..((((((((((((.......)))))....((((((.....))))))...................)))))))........(((.....))).. ( -29.50, z-score = -1.28, R) >droYak2.chr2R 4251502 94 - 21139217 CAGGUUCCACCGAGAUUUGAACUCGGAUCGCUGGAUUCAGAGUCCAGAGUGCUAACCAUUACACCAUGGAACCACCGUAUUGGUCCUGGGCCAU ..((((((((((((.......)))))....((((((.....))))))...................))))))).......((((.....)))). ( -30.70, z-score = -1.68, R) >droEre2.scaffold_4845 8826066 94 - 22589142 CAGGUUCCACCGAGAUUUGAACUCGGAUCGCUGGAUUCAGAGUCCAGAGUGCUAACCAUUACACCAUGGAACCGCCGUAUUGGUCCUGAGCCAG ..((((((((((((.......)))))....((((((.....))))))...................))))))).......((((.....)))). ( -29.40, z-score = -1.83, R) >droAna3.scaffold_13266 15704415 94 - 19884421 CAGGUUCCACCGAGAUUUGAACUCGGAUCGCUGGAUUCAGAGUCCAGAGUGCUAACCAUUACACCAUGGAACCAACGUGUUGGCCAUGAGCCAU ..((((((((((((.......)))))....((((((.....))))))...................))))))).......((((.....)))). ( -30.90, z-score = -2.42, R) >dp4.chr4_group3 8793153 90 + 11692001 AAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACC----CACUGGGCGAGGGCCAU ..(((((..(((((.......))))).((((((.((((((....)))))).))..........((((((...)----)).)))))))))))).. ( -31.30, z-score = -2.06, R) >droPer1.super_1 10244900 90 + 10282868 AAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACC----CACUGGGCGAGGGCCAU ..(((((..(((((.......))))).((((((.((((((....)))))).))..........((((((...)----)).)))))))))))).. ( -31.30, z-score = -2.06, R) >droWil1.scaffold_181096 11629861 73 + 12416693 UAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACC--------------------- ..((((((((((((.......))))).....((.((((((....)))))).)).............)))))))--------------------- ( -25.20, z-score = -3.43, R) >droVir3.scaffold_12875 13668698 94 - 20611582 CAGGUUCCACCGAGAUUUGAACUCGGAUCGCUGGAUUCAGAGUCCAGAGUGCUAACCAUUACACCAUGGAACCAACUUGCCCGGGUUGGCAAAC ..((((((((((((.......)))))....((((((.....))))))...................)))))))...(((((......))))).. ( -31.90, z-score = -2.49, R) >droMoj3.scaffold_6496 13281916 94 + 26866924 CAGGUUCCACCGAGAUUUGAACUCGGAUCGCUGGAUUCAGAGUCCAGAGUGCUAACCAUUACACCAUGGAACCAACUUGCCCGGCUUGCCAAAC ..((((((((((((.......)))))....((((((.....))))))...................))))))).........((....)).... ( -28.50, z-score = -2.22, R) >droGri2.scaffold_15112 3358604 94 - 5172618 CAGGUUCCACCGAGAUUUGAACUCGGAUCGCUGGAUUCAGAGUCCAGAGUGCUAACCAUUACACCAUGGAACCAACUUGGCCAGCUUGGCAAAA ..((((((((((((.......)))))....((((((.....))))))...................)))))))......(((.....))).... ( -31.00, z-score = -2.53, R) >anoGam1.chr2R 59258907 73 + 62725911 UAGGUUCCACCGAGAUUUGAACUCGGAUCGUUGGAUUCAGAGUCCAAAGUGCUAACCAUUACACCAUGGAACC--------------------- ..((((((((((((.......)))))....((((((.....))))))...................)))))))--------------------- ( -22.70, z-score = -2.73, R) >consensus CAGGUUCCACCGAGAUUUGAACUCGGAUCGCUGGAUUCAGAGUCCAGAGUGCUAACCAUUACACCAUGGAACCACCGUAUUGGGCCUGGGCCAU ..((((((((((((.......)))))....((((((.....))))))...................)))))))..................... (-26.22 = -25.72 + -0.50)


| Location | 12,095,138 – 12,095,232 |
|---|---|
| Length | 94 |
| Sequences | 13 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 85.31 |
| Shannon entropy | 0.33951 |
| G+C content | 0.51506 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -25.80 |
| Energy contribution | -25.31 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.871710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
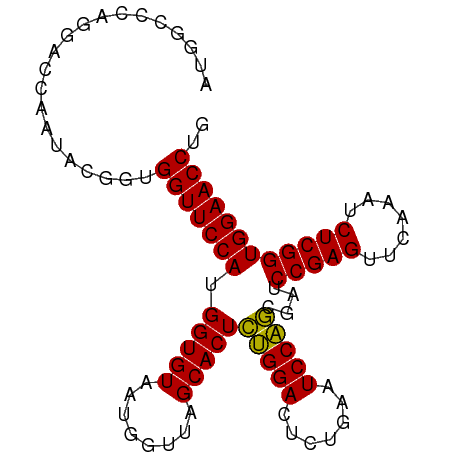
>dm3.chr2R 12095138 94 - 21146708 AUGGCCCAGGACCAAUACGGUGGUUCCAUGGUGUAAUGGUUAGCACUCUGGACUCUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUGGAACCUG .((((....).))).......((((((((.(......((((.(.((((.((((.(((....))).).))))))).).)))).).)))))))).. ( -28.50, z-score = -0.34, R) >droSim1.chr2R 10834382 94 - 19596830 AUGGCCCAGGACCAAUACGGUGGUUCCAUGGUGUAAUGGUUAGCACUCUGGACUCUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUGGAACCUG .((((....).))).......((((((((.(......((((.(.((((.((((.(((....))).).))))))).).)))).).)))))))).. ( -28.50, z-score = -0.34, R) >droSec1.super_1 9594335 94 - 14215200 AAGGCCCAGGACCAAUACGGUGGUUCCAUGGUGUAAUGGUUAGCACUCUGGACUCUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUGGAACCUG .(((.(((.(((((((((.(((....))).))))..)))))....(.(((((.......))))).)..(((((.......))))))))..))). ( -28.80, z-score = -0.52, R) >droYak2.chr2R 4251502 94 + 21139217 AUGGCCCAGGACCAAUACGGUGGUUCCAUGGUGUAAUGGUUAGCACUCUGGACUCUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUGGAACCUG .((((....).))).......((((((((.(......((((.(.((((.((((.(((....))).).))))))).).)))).).)))))))).. ( -28.50, z-score = -0.34, R) >droEre2.scaffold_4845 8826066 94 + 22589142 CUGGCUCAGGACCAAUACGGCGGUUCCAUGGUGUAAUGGUUAGCACUCUGGACUCUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUGGAACCUG .((((....).))).......((((((((.(......((((.(.((((.((((.(((....))).).))))))).).)))).).)))))))).. ( -27.50, z-score = -0.24, R) >droAna3.scaffold_13266 15704415 94 + 19884421 AUGGCUCAUGGCCAACACGUUGGUUCCAUGGUGUAAUGGUUAGCACUCUGGACUCUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUGGAACCUG .((((.....)))).......((((((((.(......((((.(.((((.((((.(((....))).).))))))).).)))).).)))))))).. ( -31.20, z-score = -1.68, R) >dp4.chr4_group3 8793153 90 - 11692001 AUGGCCCUCGCCCAGUG----GGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUU ..(((....)))....(----(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))). ( -28.90, z-score = -0.79, R) >droPer1.super_1 10244900 90 - 10282868 AUGGCCCUCGCCCAGUG----GGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUU ..(((....)))....(----(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))). ( -28.90, z-score = -0.79, R) >droWil1.scaffold_181096 11629861 73 - 12416693 ---------------------GGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUA ---------------------(((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))).. ( -25.40, z-score = -2.46, R) >droVir3.scaffold_12875 13668698 94 + 20611582 GUUUGCCAACCCGGGCAAGUUGGUUCCAUGGUGUAAUGGUUAGCACUCUGGACUCUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUGGAACCUG .((((((......))))))..((((((((.(......((((.(.((((.((((.(((....))).).))))))).).)))).).)))))))).. ( -32.20, z-score = -1.88, R) >droMoj3.scaffold_6496 13281916 94 - 26866924 GUUUGGCAAGCCGGGCAAGUUGGUUCCAUGGUGUAAUGGUUAGCACUCUGGACUCUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUGGAACCUG ((((((....)))))).....((((((((.(......((((.(.((((.((((.(((....))).).))))))).).)))).).)))))))).. ( -32.30, z-score = -1.65, R) >droGri2.scaffold_15112 3358604 94 + 5172618 UUUUGCCAAGCUGGCCAAGUUGGUUCCAUGGUGUAAUGGUUAGCACUCUGGACUCUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUGGAACCUG .....(((.(((((.....(..(.((((.(((((........))))).))))..)..)..)))))...(((((.......))))))))...... ( -29.90, z-score = -1.51, R) >anoGam1.chr2R 59258907 73 - 62725911 ---------------------GGUUCCAUGGUGUAAUGGUUAGCACUUUGGACUCUGAAUCCAACGAUCCGAGUUCAAAUCUCGGUGGAACCUA ---------------------(((((((((((((........)))))((((((((.((.((....)))).))))))))......)))))))).. ( -26.40, z-score = -3.17, R) >consensus AUGGCCCAGGACCAAUACGGUGGUUCCAUGGUGUAAUGGUUAGCACUCUGGACUCUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUGGAACCUG .....................(((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))).. (-25.80 = -25.31 + -0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:38 2011