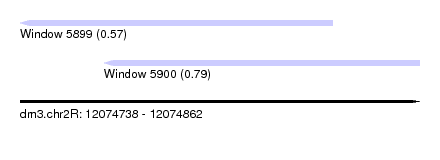
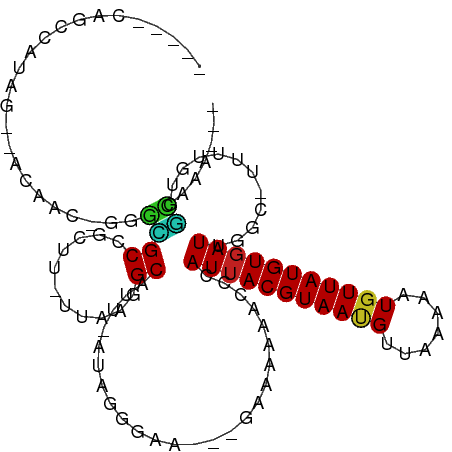
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,074,738 – 12,074,862 |
| Length | 124 |
| Max. P | 0.791683 |

| Location | 12,074,738 – 12,074,835 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.36 |
| Shannon entropy | 0.51295 |
| G+C content | 0.33422 |
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -7.10 |
| Energy contribution | -6.55 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.573187 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 12074738 97 - 21146708 AAGGCAUUAUAGGG---AAGAAAAAGCCCAUUACGUAAUGUUAAAAAUGUUAUGUGAUU-GGCUUUUGUGGCAAAUAAUAUAUUG----AGGCAGUUUCCAAGGC--------------- ............((---(((.(((((((.((((((((((((.....)))))))))))).-)))))))((..(((........)))----..))..))))).....--------------- ( -25.30, z-score = -2.51, R) >droSim1.chr2R 10812744 97 - 19596830 AAGGCAUUAUAGGG---GAGAAAAAGCCCAUUACGUAAUGUUAAAAAUGUUAUGUGAUU-GGCUUUUGUGGCAAAUAAAAUAUUG----AGGCAGUUUCCUAGGC--------------- ...((....((((.---.(..(((((((.((((((((((((.....)))))))))))).-)))))))((..(((........)))----..))..)..)))).))--------------- ( -28.20, z-score = -3.23, R) >droSec1.super_1 9574104 97 - 14215200 AAGGCAUUAUAGGG---GAGAAAAAGCCCAUUACGUAAUGUUAAAAAUGUUAUGUGAUU-GGCUUUUGUGGCAAAUAAAAUAUUG----AGGCAGUUUCCAAGGC--------------- .............(---(((((((((((.((((((((((((.....)))))))))))).-)))))))((..(((........)))----..))..))))).....--------------- ( -25.70, z-score = -2.55, R) >droYak2.chr2R 4231060 97 + 21139217 AAGGCAUAAUAGGG---GAGAAAAAGCCCAUUACGUAAUGUUAAAAAUGUUAUGUGAUU-GGCUUUUGUGGCAAAUAAAAUAUUG----AGGCAGUUUCCAAGGU--------------- .............(---(((((((((((.((((((((((((.....)))))))))))).-)))))))((..(((........)))----..))..))))).....--------------- ( -25.70, z-score = -2.87, R) >droEre2.scaffold_4845 8805557 97 + 22589142 AAGGCAUUAUAGGG---GAGAAAAAGCCCAUUACGUAAUGUUAAAAAUGUUAUGUGAUU-GGCUUUUGUGGCAAAUAAAAUAUUG----AGGCAGUUUCCAAGGU--------------- .............(---(((((((((((.((((((((((((.....)))))))))))).-)))))))((..(((........)))----..))..))))).....--------------- ( -25.70, z-score = -2.75, R) >droAna3.scaffold_13266 15682910 101 + 19884421 AAAGCAUUAUAGGGAGAAAAAAAAAACACAUUACGUAACGUUAAAAAUGUUAUGUGAUUGGGCUUUUGCGGCAAAAUAAAUAUCG----AGGCAGUUCCCAUGGA--------------- ...........(((((..........((.((((((((((((.....)))))))))))))).(((((..................)----))))..))))).....--------------- ( -20.77, z-score = -1.36, R) >dp4.chr3 4437282 90 - 19779522 --GGCAUGAUAGGG----AGAAAAAACCCAUUACGUAAUGUUAAAAAUGUUAUGUGAUU-CGAUUUUGCAGCAA---UAAUAUUG----AGGUAGUUUCCAUGG---------------- --..((((...(((----........)))((((((((((((.....)))))))))))).-.((..((((..(((---(...))))----..))))..)))))).---------------- ( -20.50, z-score = -1.86, R) >droPer1.super_2 4624962 90 - 9036312 --GGCAUGAUAGGG----AGAAAAAACCCAUUACGUAAUGUUAAAAAUGUUAUGUGAUU-CGAUUUUGCAGCAA---UAAUAUUG----AGGUAGUUUCCAUGG---------------- --..((((...(((----........)))((((((((((((.....)))))))))))).-.((..((((..(((---(...))))----..))))..)))))).---------------- ( -20.50, z-score = -1.86, R) >droVir3.scaffold_12875 4418949 108 - 20611582 AAGGCAUUAUAG------AAAAAAAAUACAUUACGUAACG-UAAAAAUGUUAUGUGAU--GGACUAUUUUUUCUCGCAAAUAUCGCACUUGACAGUU---AUUGCCAUUCGCUGGUUUUA ..((((.((.((------((((((....((((((((((((-(....))))))))))))--).....)))))))).((.......))..........)---).)))).............. ( -26.40, z-score = -2.85, R) >droMoj3.scaffold_6496 13258580 108 - 26866924 AAAGCAUUAUAG------AA---AAAUACAUUACGUAACG-UAAAAAUGUUAUGUGAU--GGACUAUUCGUUCUCGAAAAUAUCGCAUUUAACAGUUAACAUUGCCAUUUACUGGUUUUA ...((.......------..---.....((((((((((((-(....))))))))))))--).....((((....))))......)).................((((.....)))).... ( -21.70, z-score = -1.81, R) >droGri2.scaffold_15112 3336202 94 + 5172618 AAAGCAUUAUAG------AA--AAAAUACAUUACGUAACG-UAAAAAUGUUAUGUGAU--AGACUAUUUAUUUUUGUUGCUUUUGCUUUUGCUACUCGCUAAUGU--------------- ((((((..((((------((--((((((.(((((((((((-(....))))))))))))--....))))).))))))))))))).((....)).............--------------- ( -19.40, z-score = -1.72, R) >consensus AAGGCAUUAUAGGG____AGAAAAAACCCAUUACGUAAUGUUAAAAAUGUUAUGUGAUU_GGCUUUUGUGGCAAAUAAAAUAUUG____AGGCAGUUUCCAAGGC_______________ .............................(((((((((((.......)))))))))))...(((.....)))................................................ ( -7.10 = -6.55 + -0.54)

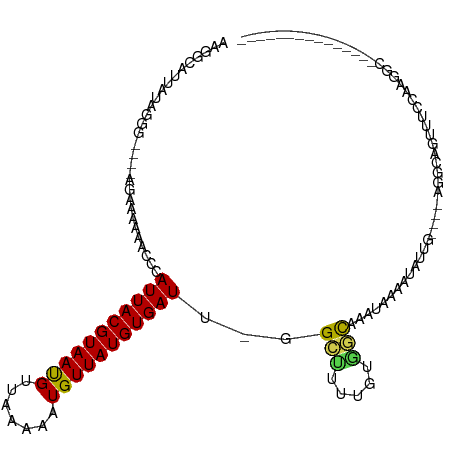
| Location | 12,074,764 – 12,074,862 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 72.44 |
| Shannon entropy | 0.55564 |
| G+C content | 0.38506 |
| Mean single sequence MFE | -25.45 |
| Consensus MFE | -8.52 |
| Energy contribution | -7.97 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.791683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12074764 98 - 21146708 -----CAGCCAUAG--ACAAC-GGGCGCCGCUU-UUAAGGCAUU-AUAGGGAA---GAAAAAGCCCAUUACGUAAUGUUAAAAAUGUUAUGUGAUUGGC-UUUUGUGGCAAA--- -----..(((((..--.....-....(((....-....)))...-........---..(((((((.((((((((((((.....)))))))))))).)))-)))))))))...--- ( -31.90, z-score = -3.06, R) >droSim1.chr2R 10812770 98 - 19596830 -----CAAGCAUAG--ACAAC-GGGUGCCGCUU-UUAAGGCAUU-AUAGGGGA---GAAAAAGCCCAUUACGUAAUGUUAAAAAUGUUAUGUGAUUGGC-UUUUGUGGCAAA--- -----...((....--.(..(-.((((((....-....))))))-...)..).---..(((((((.((((((((((((.....)))))))))))).)))-))))...))...--- ( -30.40, z-score = -2.91, R) >droSec1.super_1 9574130 98 - 14215200 -----CAAGCAUAG--ACAAC-GGGCGCCGCUU-UUAAGGCAUU-AUAGGGGA---GAAAAAGCCCAUUACGUAAUGUUAAAAAUGUUAUGUGAUUGGC-UUUUGUGGCAAA--- -----...((...(--....)-..))((((((.-....)))...-........---..(((((((.((((((((((((.....)))))))))))).)))-))))..)))...--- ( -27.70, z-score = -1.81, R) >droYak2.chr2R 4231086 98 + 21139217 -----UAGCUGUAG--ACAAC-GGGCGCCGCUU-UUAAGGCAUA-AUAGGGGA---GAAAAAGCCCAUUACGUAAUGUUAAAAAUGUUAUGUGAUUGGC-UUUUGUGGCAAA--- -----...((((..--...))-))..((((((.-....)))...-........---..(((((((.((((((((((((.....)))))))))))).)))-))))..)))...--- ( -29.10, z-score = -2.05, R) >droEre2.scaffold_4845 8805583 97 + 22589142 -----CAGCCAUAG--ACAAC--GGCGCCGCUU-UUAAGGCAUU-AUAGGGGA---GAAAAAGCCCAUUACGUAAUGUUAAAAAUGUUAUGUGAUUGGC-UUUUGUGGCAAA--- -----..(((((..--.....--...(((....-....)))...-........---..(((((((.((((((((((((.....)))))))))))).)))-)))))))))...--- ( -31.90, z-score = -3.03, R) >droAna3.scaffold_13266 15682936 106 + 19884421 AUAGCGAGCAAUAG--ACAGC-GGGCGCCGC-U-UUAAAGCAUU-AUAGGGAGAAAAAAAAAACACAUUACGUAACGUUAAAAAUGUUAUGUGAUUGGGCUUUUGCGGCAAA--- ...((..((.....--...))-..))(((((-.-..(((((...-..................((.((((((((((((.....)))))))))))))).))))).)))))...--- ( -29.75, z-score = -2.72, R) >dp4.chr3 4437304 90 - 19779522 -----GAGACAGAG--AGAAC-GGGCGCCG--------GGCAUG-AUAGGGAG----AAAAAACCCAUUACGUAAUGUUAAAAAUGUUAUGUGAUUCGA-UUUUGCAGCAAU--- -----..(.(((((--.(..(-((...)))--------..).((-(..(((..----......)))((((((((((((.....))))))))))))))).-))))))......--- ( -17.70, z-score = -0.64, R) >droPer1.super_2 4624984 90 - 9036312 -----GAGGCAGAG--AGAAC-GGGCGCCG--------GGCAUG-AUAGGGAG----AAAAAACCCAUUACGUAAUGUUAAAAAUGUUAUGUGAUUCGA-UUUUGCAGCAAU--- -----(..((((((--.(..(-((...)))--------..).((-(..(((..----......)))((((((((((((.....))))))))))))))).-))))))..)...--- ( -22.00, z-score = -1.75, R) >droWil1.scaffold_180745 2406031 100 + 2843958 ------AACCAUAGUCACACAGCAGUGCCGCUUAUAAAAGCAUUCAUAGGGAGGGAAAAAAAUAAUAUUACGUAAUGUUAAAAAUUUUAUGUUAUGUGAUUGCCAA--------- ------.....((((((((.((((...((((((....)))).(((....)))))........(((((((....))))))).........)))).))))))))....--------- ( -18.30, z-score = -0.60, R) >droVir3.scaffold_12875 4418986 96 - 20611582 -----CAGCAAUAG--ACG---GCAGGCCGCAU-UUAAGGCAUU-AUAGA------AAAAAAAUACAUUACGUAACG-UAAAAAUGUUAUGUGAUGGACUAUUUUUUCUCGCAAA -----..((.....--...---....(((....-....)))...-..(((------(((((....((((((((((((-(....))))))))))))).....)))))))).))... ( -26.60, z-score = -4.12, R) >droMoj3.scaffold_6496 13258620 94 - 26866924 -----CAGCAAUAA--ACGA--GCAGGCCGCAU-UUAAAGCAUU-AUAG---------AAAAAUACAUUACGUAACG-UAAAAAUGUUAUGUGAUGGACUAUUCGUUCUCGAAAA -----........(--((((--(.((...((..-.....))...-....---------.......((((((((((((-(....)))))))))))))..)).))))))........ ( -21.40, z-score = -2.53, R) >droGri2.scaffold_15112 3336227 97 + 5172618 -----CAGCAAUAG--ACGACGACGGGCCGCAU-UUAAAGCAUU-AUAG--------AAAAAAUACAUUACGUAACG-UAAAAAUGUUAUGUGAUAGACUAUUUAUUUUUGUUGC -----..(((((((--(....((..(...((..-.....))...-....--------.........(((((((((((-(....))))))))))))...)..))....)))))))) ( -18.60, z-score = -1.25, R) >consensus _____CAGCCAUAG__ACAAC_GGGCGCCGCUU_UUAAGGCAUU_AUAGGGAA___GAAAAAACCCAUUACGUAAUGUUAAAAAUGUUAUGUGAUUGGC_UUUUGUGGCAAA___ ........................((((...........)).........................(((((((((((.......)))))))))))............))...... ( -8.52 = -7.97 + -0.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:37 2011