| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,015,724 – 3,015,815 |
| Length | 91 |
| Max. P | 0.998550 |

| Location | 3,015,724 – 3,015,815 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 59.64 |
| Shannon entropy | 0.78214 |
| G+C content | 0.34954 |
| Mean single sequence MFE | -20.76 |
| Consensus MFE | -6.67 |
| Energy contribution | -6.97 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.80 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.40 |
| SVM RNA-class probability | 0.998550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
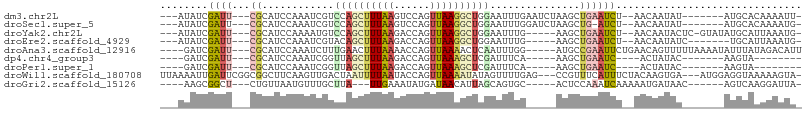
>dm3.chr2L 3015724 91 - 23011544 ---AUAUCGAUU---CGCAUCCAAAUCGUCCAGCUUUAAGUCCAGUUAAGGCUGGAAUUUGAAUCUAAGCUGAAUCU--AACAAUAU-------AUGCACAAAAUU- ---.....((((---(((((.(((((..(((((((((((......)))))))))))))))).))....)).))))).--........-------............- ( -21.30, z-score = -3.06, R) >droSec1.super_5 1172730 90 - 5866729 ---AUAUCGAUU---CGCAUCCAAAUCGUCCAGCUUUAAGUCCAGUUAAGGCUGGAAUUUGGAUCUAAGCUG-AUCU--AACAAUAU-------AUGCACAAAAUG- ---.....((((---.((((((((((..(((((((((((......)))))))))))))))))))....)).)-))).--........-------............- ( -26.40, z-score = -4.23, R) >droYak2.chr2L 3010953 92 - 22324452 ---AUAUCGAUU---CGCAUCCAAAAUGUCCAGCUUUAAGACCAGUUAAGGCUGGAAUUUG-----AAGCUGAAUCU--AACAAUACUC-GUAUAUGCAUUAAAUG- ---.....((((---(((...((((...(((((((((((......))))))))))).))))-----..)).))))).--..........-................- ( -22.80, z-score = -3.02, R) >droEre2.scaffold_4929 3062265 86 - 26641161 ---AUAUCGAUU---CGCAUCCAAAUCGUACAGCUUUAAGACCAGUUAAGGCUGGAAUUUG-----AAGCUGAAUCU--AACAAUAUC-------UGCAUUAAAUG- ---....(((((---........)))))..((((((((((.(((((....)))))..))))-----)))))).....--.........-------...........- ( -19.90, z-score = -2.34, R) >droAna3.scaffold_12916 4438564 95 - 16180835 ----GAUCGAUU---CGCAUCCAAAUCUUUGAACUUUAAAACCAGUUAAAACUCAAUUUGG-----AUGCCGAAUUCUGAACAGUUUUUAAAAUAUUUAUAGACAUU ----....((((---(((((((((((...(((..(((((......)))))..)))))))))-----))).)))))).......((((.(((.....))).))))... ( -19.60, z-score = -3.24, R) >dp4.chr4_group3 4435656 76 + 11692001 ----GAUCGAUU---CGCAUCCAAAUCGGUUAGCUUUAAGACCAGUUAAAGCUCGAUUUCA-----AAGCUGAAUC----ACUAUAC-------AAGUA-------- ----....((((---(((....((((((...((((((((......))))))))))))))..-----..)).)))))----.......-------.....-------- ( -18.20, z-score = -3.04, R) >droPer1.super_1 1581007 76 + 10282868 ----GAUCGAUU---CGCAUCCAAAUCGGUUAGCUUUAAGACCAGUUAAAGCUCGAUUUCA-----AAGCUGAAUC----ACUAUAC-------AAGUA-------- ----....((((---(((....((((((...((((((((......))))))))))))))..-----..)).)))))----.......-------.....-------- ( -18.20, z-score = -3.04, R) >droWil1.scaffold_180708 9842400 100 + 12563649 UUAAAAUUGAUUCGGCGGCUUCAAGUUGACUAAUUUUAAUACCAGUUAAAAUAUAGUUUUGAG---CCGUUUCAUUUCUACAAGUGA---AUGGAGGUAAAAAGUA- ......(((.(((.((((((.(((...(((((((((((((....)))))))).))))))))))---))))(((((((....))))))---)..))).)))......- ( -21.40, z-score = -1.81, R) >droGri2.scaffold_15126 6682519 85 - 8399593 ----AAGCGGCU---CUGUUAAUGUUUGCUUA---UUGAAAUAUGAUAACAUUAGCAGUGC-----ACUCCAAAUCAAAAAUGAUAAC------AGUCAAGGAUUA- ----.....((.---(((((((((((...(((---(......)))).))))))))))).))-----......((((.....((((...------.))))..)))).- ( -19.00, z-score = -1.70, R) >consensus ___AUAUCGAUU___CGCAUCCAAAUCGUCCAGCUUUAAGACCAGUUAAAGCUGGAAUUUG_____AAGCUGAAUCU__AACAAUAU_______AUGCAUAAAAUG_ .............................((((((((((......)))))))))).................................................... ( -6.67 = -6.97 + 0.30)
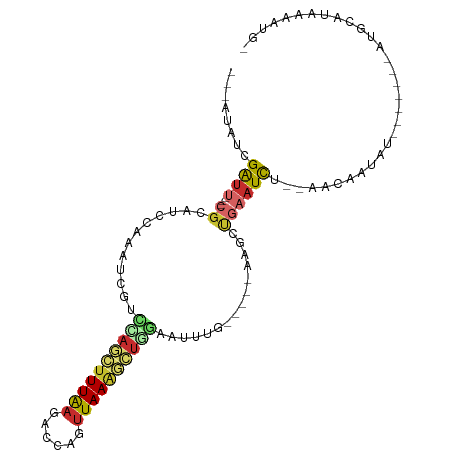
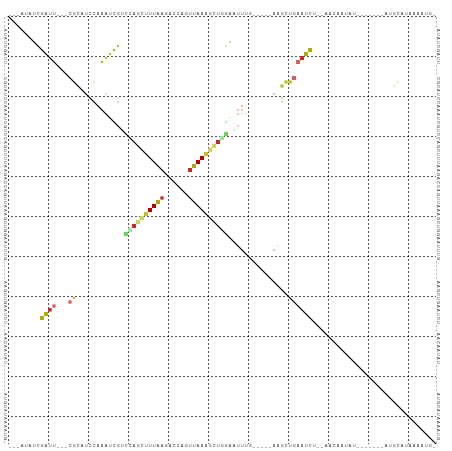
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:12:47 2011