| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,012,902 – 12,013,000 |
| Length | 98 |
| Max. P | 0.581711 |

| Location | 12,012,902 – 12,013,000 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 65.06 |
| Shannon entropy | 0.70633 |
| G+C content | 0.64810 |
| Mean single sequence MFE | -41.09 |
| Consensus MFE | -12.34 |
| Energy contribution | -12.35 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.581711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
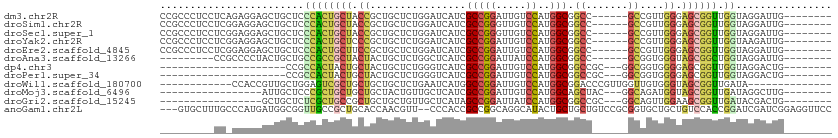
>dm3.chr2R 12012902 98 - 21146708 CCGCCCUCCUCAGAGGAGCUGCUCCCACUGCUACCGCUGCUCUGGAUCAUCGCCGGAUUGUCCAUGGCGGCC------GCCGUUGGGAGCGGUUGGUAGGAUUG-------- ((..((((....))))((((((((((((.((....((((((.(((((.(((....))).))))).)))))).------)).).)))))))))))....))....-------- ( -45.90, z-score = -2.17, R) >droSim1.chr2R 10750489 98 - 19596830 CCGCCCUCCUCGGAGGAGCUGCUCCCACUGCUACCGCUGCUCUGGAUCAUCGCCGGGUUGUCCAUGGCGGCC------GCCGUUGGGAGCGGUUGGUAGGAUUG-------- ((..((((....))))((((((((((((.((....((((((.(((((.(((....))).))))).)))))).------)).).)))))))))))....))....-------- ( -47.20, z-score = -1.73, R) >droSec1.super_1 9508244 98 - 14215200 CCGCCCUCCUCGGAGGAGCUGCUCCCACUGCUACCGCUGCUCUGGAUCAUCGCCGGGUUGUCCAUGGCGGCC------GCCGUUGGGAGCGGUUGGUAGGAUUG-------- ((..((((....))))((((((((((((.((....((((((.(((((.(((....))).))))).)))))).------)).).)))))))))))....))....-------- ( -47.20, z-score = -1.73, R) >droYak2.chr2R 4168369 98 + 21139217 CCGCCCUCCUCGGAGGAGCUGCUCCCACUGCUCCCGCUGCUCUGGAUCAUCGCCGGAUUGUCCAUGGCGGCC------GCCGUUGGGAGCGGUUGGUAAGAUUG-------- ((..((((....))))((((((((((((.((....((((((.(((((.(((....))).))))).)))))).------)).).)))))))))))))........-------- ( -47.10, z-score = -2.44, R) >droEre2.scaffold_4845 8744176 98 + 22589142 CCGCCCUCCUCGGAGGAGCUGCUCCCACUGCUUCCGCUGCUCUGGAUCAUCGCCGGAUUGUCCAUGGCGGCC------GCCGUUGGGAGCGGUUGGUAGGAUUG-------- ((..((((....))))((((((((((((.((....((((((.(((((.(((....))).))))).)))))).------)).).)))))))))))....))....-------- ( -47.80, z-score = -2.30, R) >droAna3.scaffold_13266 4488681 89 + 19884421 ---------CCGCCCCUACUGCUGCCGCCGCUACUACUGCUCUGGCUCAUCGCCGGAUUAUCCAUGGCGGCC------GCGGUGGGUAGCGGCUGGUAGGAUUG-------- ---------((((.((((((((.((((((((.......))((((((.....))))))........)))))).------))))))))..))))............-------- ( -44.60, z-score = -2.58, R) >dp4.chr3 18633959 80 - 19779522 ---------------------CCGCCACUACUGCUACUGCUCUGGGUCAUCGCCGGAUUGUCCAUGGCGGCCGC---GGCGGUGGGGAGCGGUUGGUAGGACUG-------- ---------------------.........(((((((((((((...((((((((((..(((.....)))..).)---))))))))))))))).)))))).....-------- ( -35.30, z-score = -0.78, R) >droPer1.super_34 581012 80 - 916997 ---------------------CCGCCACUACUGCUACUGCUCUGGGUCAUCGCCGGAUUGUCCAUGGCGGCCGC---GGCGGUGGGGAGCGGUUGGUAGGACUG-------- ---------------------.........(((((((((((((...((((((((((..(((.....)))..).)---))))))))))))))).)))))).....-------- ( -35.30, z-score = -0.78, R) >droWil1.scaffold_180700 4227312 86 - 6630534 ------------CCACCGUUGCUGGAGUCGCUGCUGCUGCUCUGAAUCAUGGCCGGAUUGUCCAUGGCGGACCCGUUGGUUGUGGGUAGCGGUUGAUA-------------- ------------(.((((((((((((((.((....)).)))))....((..(((((...((((.....))))...)))))..))))))))))).)...-------------- ( -36.70, z-score = -2.19, R) >droMoj3.scaffold_6496 16342037 84 + 26866924 -----------------AUUGCUCCCGCUGCUGCUGCUACUGUUGCUCAUCGCCGGAUUGUCCAUGGCAGCUAC---GGCAGAUGGUAGCGGUUGAUAGGCUUG-------- -----------------...(((.((((((((((((((...(((((.(((....((.....))))))))))...---))))).)))))))))......)))...-------- ( -31.20, z-score = -0.73, R) >droGri2.scaffold_15245 16993060 84 - 18325388 -----------------GCUGCUCUCGCUGCCGCUGCUGCUGUUGCUCAUAGCCGGAUUAUCCAUGGCGGCCGC---GGCAGUUGGAAGCGGUUGAUACGACUG-------- -----------------((((((((.((((((((.((((((((.((.....)).((.....)))))))))).))---)))))).)).))))))...........-------- ( -39.80, z-score = -2.79, R) >anoGam1.chr2L 19211883 107 + 48795086 ---GUGCUUUGCCCAUGAUGGCGGUUGCCGCUGCACCAACGUU--CCCACCGCCGGCAGGCAUACUGCUGCUGUCCGCGGUGCUGCUGUCCACCGGAUCGAUCGGAGGUUCC ---.......(((...(((((((((.((....)))))......--..((((((.(((((((.....))..))))).))))))..))))))..((((.....)))).)))... ( -35.00, z-score = 1.81, R) >consensus _________UC_GAGGAGCUGCUCCCACUGCUGCUGCUGCUCUGGAUCAUCGCCGGAUUGUCCAUGGCGGCC______GCCGUUGGGAGCGGUUGGUAGGAUUG________ ........................((((((((.((................(((((.....))..))).((.......))....)).)))))).))................ (-12.34 = -12.35 + 0.01)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:30 2011