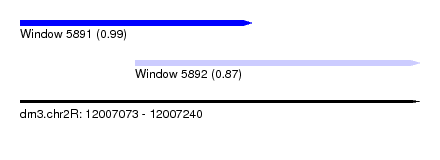
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,007,073 – 12,007,240 |
| Length | 167 |
| Max. P | 0.994133 |

| Location | 12,007,073 – 12,007,170 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.90 |
| Shannon entropy | 0.37506 |
| G+C content | 0.34006 |
| Mean single sequence MFE | -31.03 |
| Consensus MFE | -22.90 |
| Energy contribution | -22.15 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.994133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
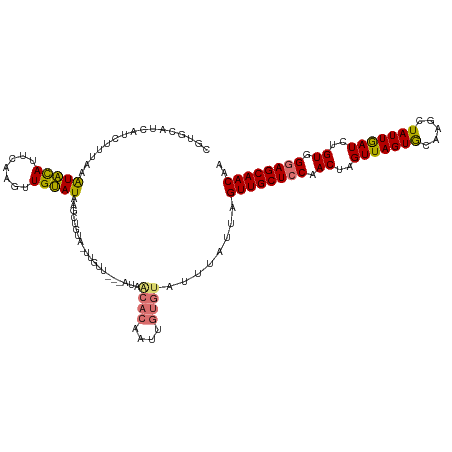
>dm3.chr2R 12007073 97 + 21146708 CGUGCAUCAUCUUUGAAUACAUUCAAGUUGUAUAAGCUGUA-CUAUU---AU-------------------UAGUUGCUUCAACUAGUUAGUACAAGCUAUUGAUCUGUGGUAGCAACAA ..((((((((..(..((((((.......))))).(((((((-(((..---((-------------------((((((...)))))))))))))).)))).)..)...))))).))).... ( -25.60, z-score = -2.12, R) >droSim1.chr2R 10744810 117 + 19596830 CGUGCAUCAUCUUGAAAUACAUUCAAGUUGUAUAAGCUGCAAUUGUU---AUAACACAAUUGUGUAUUUAUUAGUUGCUCCAACUAGUUAGUGCAAGCUAUUGAUCUGUGGGAGCAACAA .(((((....((((((.....)))))).)))))..((.((((((((.---.....))))))))))........((((((((.((..(((((((.....)))))))..)).)))))))).. ( -35.10, z-score = -3.03, R) >droSec1.super_1 9502600 116 + 14215200 CGUGCAUCAUCUUUAAAUACAUUCAAGUUGUAUAAGCUGUG-UUGUU---AUAACACAAUUGUGUAUUUAUUAGUUGCUCCAACUAGUUAGUGCUAGCUAUUGAUCUGUGGGAGCAACAA .............(((((((((...((((.....))))(((-(((..---.))))))....)))))))))...((((((((.((..(((((((.....)))))))..)).)))))))).. ( -32.10, z-score = -2.32, R) >droYak2.chr2R 4162520 117 - 21139217 ---CUCUAGUAUUUAAGUGUAUUCAAGUUGCAUAAGAUAUAAUUGUUGUAAUGGCACAAUUGUGUAUCCAAUAGUUGCUCCAACUAGUUAGUGAAAACUAUUAAUCUGUGGGAGCAACAA ---.............(((((.......)))))...((((((((((.((....))))))))))))........((((((((.((..(((((((.....)))))))..)).)))))))).. ( -31.30, z-score = -2.89, R) >consensus CGUGCAUCAUCUUUAAAUACAUUCAAGUUGUAUAAGCUGUA_UUGUU___AUAACACAAUUGUGUAUUUAUUAGUUGCUCCAACUAGUUAGUGCAAGCUAUUGAUCUGUGGGAGCAACAA ................(((((.......)))))....................((((....))))........((((((((.((..(((((((.....)))))))..)).)))))))).. (-22.90 = -22.15 + -0.75)

| Location | 12,007,121 – 12,007,240 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.94 |
| Shannon entropy | 0.21689 |
| G+C content | 0.36994 |
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -25.67 |
| Energy contribution | -25.18 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.867675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
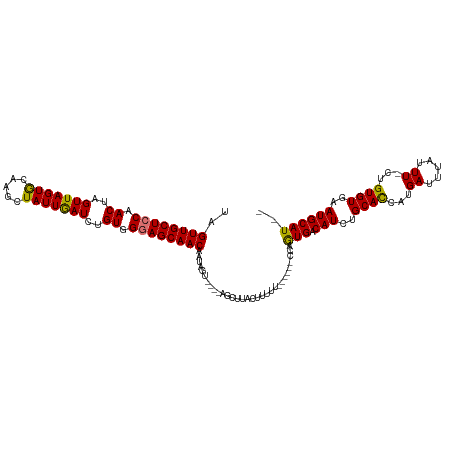
>dm3.chr2R 12007121 119 + 21146708 UAGUUGCUUCAACUAGUUAGUACAAGCUAUUGAUCUGUGGUAGCAACAAUAGUUAGUAACUUACUUUUUUUUUUGCCAAUGACAUCUGCACCAUGAUUUAUUU-CCGUGUGAAUGCAUCC ((((((...))))))((.(((((.((((((((...(((....))).)))))))).)).))).))..............(((.(((..((((...((......)-).))))..)))))).. ( -22.20, z-score = -0.02, R) >droSim1.chr2R 10744878 108 + 19596830 UAGUUGCUCCAACUAGUUAGUGCAAGCUAUUGAUCUGUGGGAGCAACAAUAGU----AGCUUACUUUUU------CCAGUGACAUCUGCACCAUGAUUUAUUUUCUGUGUGAAUGCAU-- ..((((((((.((..(((((((.....)))))))..)).))))))))......----............------...(((.(((..((((...((.......)).))))..))))))-- ( -27.70, z-score = -0.84, R) >droSec1.super_1 9502667 108 + 14215200 UAGUUGCUCCAACUAGUUAGUGCUAGCUAUUGAUCUGUGGGAGCAACAAUAGU----AGCUUACUUUUU------CCAGUGACAUCUGCACCAUGAUUUAUUUUCUGUGUGAAUGCAU-- ..((((((((.((..(((((((.....)))))))..)).))))))))......----............------...(((.(((..((((...((.......)).))))..))))))-- ( -27.70, z-score = -0.83, R) >droYak2.chr2R 4162588 106 - 21139217 UAGUUGCUCCAACUAGUUAGUGAAAACUAUUAAUCUGUGGGAGCAACAA-AAU----ACCAUACCUUUU------UAAGUGACAUCUGCAUCGUGAUUUAUUU-CUGUGUGAAUGCAU-- ..((((((((.((..(((((((.....)))))))..)).))))))))..-...----..(((((.....------(((((.((.........)).)))))...-..))))).......-- ( -27.70, z-score = -2.75, R) >consensus UAGUUGCUCCAACUAGUUAGUGCAAGCUAUUGAUCUGUGGGAGCAACAAUAGU____AGCUUACUUUUU______CCAGUGACAUCUGCACCAUGAUUUAUUU_CUGUGUGAAUGCAU__ ..((((((((.((..(((((((.....)))))))..)).))))))))...............................(((.(((..((((...((.....))...))))..)))))).. (-25.67 = -25.18 + -0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:29 2011