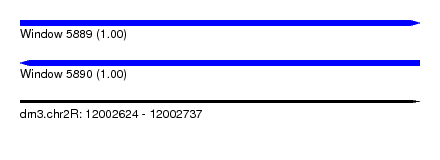
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,002,624 – 12,002,737 |
| Length | 113 |
| Max. P | 0.999056 |

| Location | 12,002,624 – 12,002,737 |
|---|---|
| Length | 113 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 63.11 |
| Shannon entropy | 0.73318 |
| G+C content | 0.51990 |
| Mean single sequence MFE | -37.56 |
| Consensus MFE | -11.65 |
| Energy contribution | -11.24 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.46 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
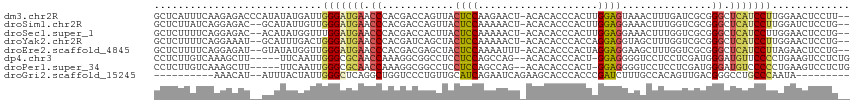
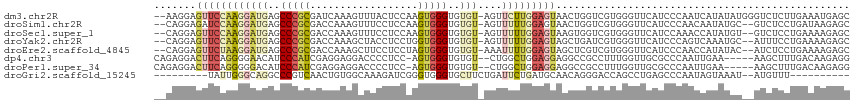
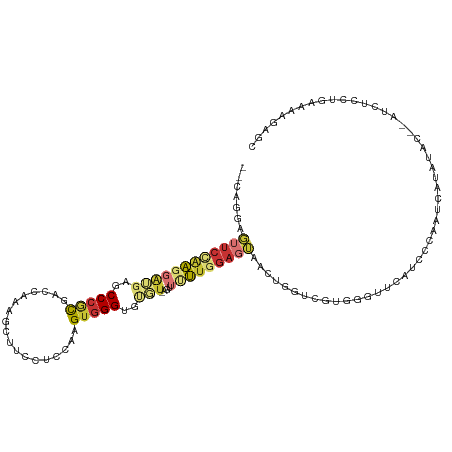
>dm3.chr2R 12002624 113 + 21146708 GCUCAUUUCAAGAGACCCAUAUAUGAUUGGGAUGAACCCACGACCAGUUACUCCAAGAACU-ACACACCCACUUGGAGUAAACUUUGAUCGCGGGCUCAUCCUUGGAACUCCUU-- .....(((((((.((((((........)))).(((.(((.(((.(((((((((((((....-.........))))))))))...))).))).))).))))))))))))......-- ( -37.12, z-score = -4.89, R) >droSim1.chr2R 10740122 111 + 19596830 GCUCUUAUCAGGAGAC--GCAUAUUGUUGGGAUGAACCCACGACCAGUUACUCCAAAAACU-ACACACCCACUUGGAGGAAACUUUGGUCGCGGGCUCAUCCUUGGAUCUCCUG-- ........(((((((.--.((.......(((((((.(((.(((((((((.((((((.....-..........))))))..)))..)))))).))).)))))))))..)))))))-- ( -43.57, z-score = -5.33, R) >droSec1.super_1 9498116 111 + 14215200 GCUCUUUUCAGGAGAC--ACAUAUGGUUUGGAUGAACCCACGACCACUUACUCCAAAAACU-ACACACCCACUUGGAGGAAACUUUGGUCGCGGGCUCAUCCUUGGAACUCCUG-- ........((((((.(--(....))....((((((.(((.((((((....((((((.....-..........)))))).......)))))).))).))))))......))))))-- ( -38.96, z-score = -5.04, R) >droYak2.chr2R 4158087 111 - 21139217 GCUCUUUUCAGGAAAU--GCAUUUGACUGGGAUGAACCCACGAUCAGCUACUCCAAAAACU-ACACACCCACCAGGAGGUAGCUUUGGUCGCGGGCUCAUCCUUGGAACUCCUG-- ........(((((..(--.((.......(((((((.(((.((((((((((((((.......-............))).)))))..)))))).))).))))))))).)..)))))-- ( -38.02, z-score = -3.33, R) >droEre2.scaffold_4845 8734110 111 - 22589142 GCUCUUUUCAGGAGAU--GUAUAUGGUUGGGAUGAACCCACGACGAGCUACUCCAAAAUUU-ACACACCCACUAGGAGGAAGCUUUGGUCGCGGGCUCAUCCUUAGAACUCCUG-- ........((((((.(--.((.......(((((((.(((.(((((((((.((((.......-............))))..)))))..)))).))).))))))))).).))))))-- ( -39.32, z-score = -3.46, R) >dp4.chr3 18619673 108 + 19779522 CCUCUUGUCAAAGCUU-----UUCAAUUGGGCGCAACCAAAGGCGGCCUCCUCCAGCCAG--ACACACCCACU-GGAGGGGUCCUCCUCGAUGGGAUGUUCCCCUGAAGUCCUCUG ........((.((..(-----((((...(((.(((.(((.(((.((.(((((((((....--.........))-))))))).)).)))...)))..))).))).)))))..)).)) ( -35.52, z-score = -0.57, R) >droPer1.super_34 566149 108 + 916997 CCUCUUGUCAAAGCUU-----UUCAAUUGGGCGCAACCAAAGGCGGCCUCCUCCAGCCAG--ACACACCCACU-GGAGGGGUCCUCCUCGAUGGGAUGUCCCCCUGAAGUCCUCUG ........((.((..(-----((((...(((.(((.(((.(((.((.(((((((((....--.........))-))))))).)).)))...)))..))).))).)))))..)).)) ( -35.52, z-score = -0.48, R) >droGri2.scaffold_15245 16979493 95 + 18325388 ----------AAACAU--AUUUACUAUUGGGCUCAGGCUGGUCCCUGUUGCAUCAGAAUCAGAAGCACCCACCCGAUCUUUGCCACAGUUGACGGGCCUGCCCAAUA--------- ----------......--......((((((((..(((((.(((.((((.(((..(((.((..............))))).))).))))..))).)))))))))))))--------- ( -32.44, z-score = -2.80, R) >consensus GCUCUUUUCAAGAGAU__ACAUAUGAUUGGGAUGAACCCACGACCAGCUACUCCAAAAACU_ACACACCCACUUGGAGGAAACUUUGGUCGCGGGCUCAUCCUUGGAACUCCUG__ ............................(((((((.((............((((....................))))...............)).)))))))............. (-11.65 = -11.24 + -0.41)
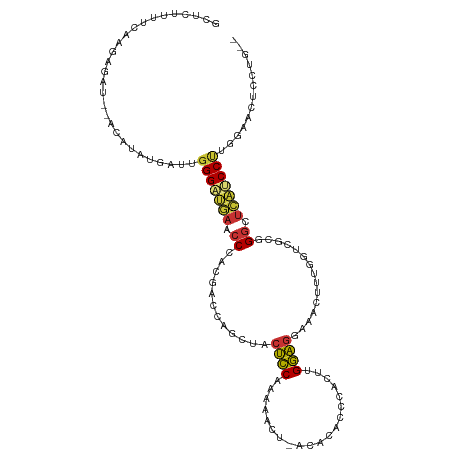
| Location | 12,002,624 – 12,002,737 |
|---|---|
| Length | 113 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 63.11 |
| Shannon entropy | 0.73318 |
| G+C content | 0.51990 |
| Mean single sequence MFE | -45.25 |
| Consensus MFE | -7.58 |
| Energy contribution | -8.14 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.59 |
| Mean z-score | -4.07 |
| Structure conservation index | 0.17 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.23 |
| SVM RNA-class probability | 0.998007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12002624 113 - 21146708 --AAGGAGUUCCAAGGAUGAGCCCGCGAUCAAAGUUUACUCCAAGUGGGUGUGU-AGUUCUUGGAGUAACUGGUCGUGGGUUCAUCCCAAUCAUAUAUGGGUCUCUUGAAAUGAGC --.(((((..(((.(((((((((((((((((....((((((((((.........-....)))))))))).)))))))))))))))))..........)))..)))))......... ( -49.32, z-score = -6.05, R) >droSim1.chr2R 10740122 111 - 19596830 --CAGGAGAUCCAAGGAUGAGCCCGCGACCAAAGUUUCCUCCAAGUGGGUGUGU-AGUUUUUGGAGUAACUGGUCGUGGGUUCAUCCCAACAAUAUGC--GUCUCCUGAUAAGAGC --((((((((.((.(((((((((((((((((..(((..(((((((.........-....))))))).))))))))))))))))))))........)).--))))))))........ ( -52.12, z-score = -6.71, R) >droSec1.super_1 9498116 111 - 14215200 --CAGGAGUUCCAAGGAUGAGCCCGCGACCAAAGUUUCCUCCAAGUGGGUGUGU-AGUUUUUGGAGUAAGUGGUCGUGGGUUCAUCCAAACCAUAUGU--GUCUCCUGAAAAGAGC --((((((......(((((((((((((((((.......(((((((.........-....)))))))....)))))))))))))))))....((....)--).))))))........ ( -48.42, z-score = -6.55, R) >droYak2.chr2R 4158087 111 + 21139217 --CAGGAGUUCCAAGGAUGAGCCCGCGACCAAAGCUACCUCCUGGUGGGUGUGU-AGUUUUUGGAGUAGCUGAUCGUGGGUUCAUCCCAGUCAAAUGC--AUUUCCUGAAAAGAGC --((((((......((((((((((((((....((((((.(((............-.......)))))))))..))))))))))))))..((.....))--..))))))........ ( -43.41, z-score = -3.61, R) >droEre2.scaffold_4845 8734110 111 + 22589142 --CAGGAGUUCUAAGGAUGAGCCCGCGACCAAAGCUUCCUCCUAGUGGGUGUGU-AAAUUUUGGAGUAGCUCGUCGUGGGUUCAUCCCAACCAUAUAC--AUCUCCUGAAAAGAGC --((((((......(((((((((((((((...((((..((((....((((....-..)))).)))).)))).)))))))))))))))...........--..))))))........ ( -44.81, z-score = -4.92, R) >dp4.chr3 18619673 108 - 19779522 CAGAGGACUUCAGGGGAACAUCCCAUCGAGGAGGACCCCUCC-AGUGGGUGUGU--CUGGCUGGAGGAGGCCGCCUUUGGUUGCGCCCAAUUGAA-----AAGCUUUGACAAGAGG (((((...((((.(((..((..(((..((((.((.(((((((-(((.((.....--)).)))))))).)))).))))))).))..)))...))))-----...)))))........ ( -44.20, z-score = -1.45, R) >droPer1.super_34 566149 108 - 916997 CAGAGGACUUCAGGGGGACAUCCCAUCGAGGAGGACCCCUCC-AGUGGGUGUGU--CUGGCUGGAGGAGGCCGCCUUUGGUUGCGCCCAAUUGAA-----AAGCUUUGACAAGAGG (((((...(((((((.(.((..(((..((((.((.(((((((-(((.((.....--)).)))))))).)))).))))))).))).)))...))))-----...)))))........ ( -44.00, z-score = -1.10, R) >droGri2.scaffold_15245 16979493 95 - 18325388 ---------UAUUGGGCAGGCCCGUCAACUGUGGCAAAGAUCGGGUGGGUGCUUCUGAUUCUGAUGCAACAGGGACCAGCCUGAGCCCAAUAGUAAAU--AUGUUU---------- ---------((((((((((((..(((..((((.(((.((((((((........))))).)))..))).)))).)))..))))..))))))))......--......---------- ( -35.70, z-score = -2.15, R) >consensus __CAGGAGUUCCAAGGAUGAGCCCGCGACCAAAGCUUCCUCCAAGUGGGUGUGU_AGUUUUUGGAGUAACUGGUCGUGGGUUCAUCCCAAUCAUAUAC__AUCUCCUGAAAAGAGC .......((((((((((((..(((((..................)))))..)))....)))))))))................................................. ( -7.58 = -8.14 + 0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:27 2011