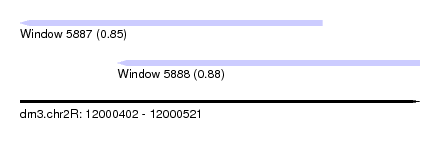
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,000,402 – 12,000,521 |
| Length | 119 |
| Max. P | 0.884527 |

| Location | 12,000,402 – 12,000,492 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 57.81 |
| Shannon entropy | 0.87444 |
| G+C content | 0.52648 |
| Mean single sequence MFE | -23.71 |
| Consensus MFE | -8.52 |
| Energy contribution | -8.21 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 2.44 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.854026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
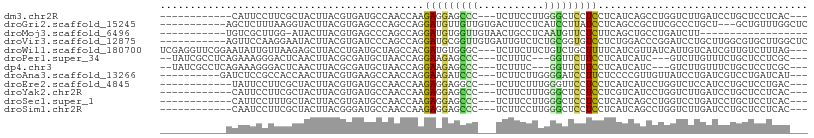
>dm3.chr2R 12000402 90 - 21146708 ------------CAUUCCUUCGCUACUUACGUGAUGCCAACCAAGAGGAGCCC---UCUUCCUUGGGCUCCUCCUCAUCAGCCUGGUCUUGAUCCUGCUCCUCAC--- ------------..................((((..........(((((((((---........)))))))))......(((..((.......)).)))..))))--- ( -25.30, z-score = -2.31, R) >droGri2.scaffold_15245 16976923 94 - 18325388 -----------AGCUCUUUAAGGUACUUACGUGAGCCCAGCCAGGAUGUUGUUGUGACUUCCUCAUCCUUAUCCUCAGCCGCUUCGCCCUGCU---GCUGUUUGGCUC -----------.((((..((((...))))...))))..((((((((((.....((((.....))))...))))).((((.((........)).---))))..))))). ( -25.30, z-score = -0.96, R) >droMoj3.scaffold_6496 16322860 78 + 26866924 -----------UGUCGCUUGG-AUACUUACGUGAGCCCAGCCAGGAUGUGGUUGUAACUGCCUCAAUGUUCUCUUCAGCUGCCUGAUCUU------------------ -----------....(((.((-.(((....)))..)).)))((((....(((((.....((......))......))))).)))).....------------------ ( -14.20, z-score = 1.67, R) >droVir3.scaffold_12875 17464425 97 - 20611582 -----------AGUUCCAAGGAAUACUUACGUGAUCCCAGCCAGGAUGCGGUUGUGAUUGUCUCUGCGGUGUCCUCUGGACCCGGAUCCUGCUUGGCGUGCUUGGCUC -----------....(((((((.(((....))).))((((.((((((.(((.....(((((....)))))((((...))))))).)))))).))))....)))))... ( -29.70, z-score = 0.21, R) >droWil1.scaffold_180700 4164267 102 - 6630534 UCGAGGUUCGGAAUAUUGUUAAGAGCUUACCUGAUGCUAGCCACGAUGUGGGC---UCUUCUUCUGUCUGCUUUUCAUCGUUAUCAUUGUCAUCGUUGUCUUUAG--- ...((((((..(((...)))..))))))....(((((.....((((((.((((---.............))))..)))))).......).))))...........--- ( -17.52, z-score = 1.45, R) >droPer1.super_34 563906 94 - 916997 --UAUCGCCUCAGAAAGGGACUCAACUUACGCGAUGCUAACCAGGAAGAGCCC---UCUUUC---GGUUCUUCCUCAUCAUC---GUCUUGUUUCUGCUCCUCGC--- --.............(((..(..(((....((((((......((((((((((.---......---))))))))))...))))---))...)))...)..)))...--- ( -25.40, z-score = -1.72, R) >dp4.chr3 18617424 94 - 19779522 --UAUCGCCUCAGAAAGGGACUCAACUUACGCGAUGCUAACCAGGAAGAGCCC---UCUUUC---GGUUCUUCCUCAUCAUC---GUCUUGUUUCUGCUCCUCGC--- --.............(((..(..(((....((((((......((((((((((.---......---))))))))))...))))---))...)))...)..)))...--- ( -25.40, z-score = -1.72, R) >droAna3.scaffold_13266 4477187 92 + 19884421 ----------GAUCUCCGCCACCAACUUACGUGAAGCCAACCAGGAAGAUCCC---UCUUCUUGGGGAUCCUUCUCCCCGUUGUUAUCCUGAUCGUCCUGAUCAU--- ----------((((...(((((........)))..)).....((((.((((..---......((((((......))))))..........)))).))))))))..--- ( -23.07, z-score = -1.35, R) >droEre2.scaffold_4845 8731916 90 + 22589142 ------------UAUUCCUUCGCUACUUACGUGAUGCCAACCAAGAGGAGGCC---UCUUCUUUGGGUUCCUCCUCAUCAUCCUGGUCUCCAUCCUGCUCCUGAC--- ------------..................(.((.((((.....(((((((.(---((......)))..))))))).......)))).)))..............--- ( -19.30, z-score = -0.21, R) >droYak2.chr2R 4155876 90 + 21139217 ------------CAUUCCUUCGCUACUUACGUGAUGCCAACCAAGAGGAGCCC---UCUUCUUUGGGCUCCUCCUCGUCAUCCUGGUCUUGAUCCUGCUCCUCAC--- ------------.......(((..(((...((((((........(((((((((---........)))))))))..))))))...)))..))).............--- ( -25.40, z-score = -2.83, R) >droSec1.super_1 9495914 90 - 14215200 ------------CAUUCCUUUGCUACUUACGUGAUGCCAACCAAGAGGAGCCC---UCUUCCUUGGGCUCCUCCUCAUCAGCCUGGUCCUGAUCCUGCUCCUCAC--- ------------..................((((.((.......(((((((((---........)))))))))...(((((.......)))))...))...))))--- ( -26.00, z-score = -2.45, R) >droSim1.chr2R 10737907 90 - 19596830 ------------CAAUCCUUCGCUACUUACGGGAUGCCAACCAAGAGGAGCCC---UCUUCCUUGGGCUCCUCCUCAUCAGCCUGGUCUUGAUCCUGCUCCUCAC--- ------------.................((((((....((((.(((((((((---........)))))))))((....))..))))....))))))........--- ( -27.90, z-score = -2.23, R) >consensus ___________AGAUUCCUUCGCUACUUACGUGAUGCCAACCAGGAGGAGCCC___UCUUCCUUGGGCUCCUCCUCAUCAUCCUGGUCUUGAUCCUGCUCCUCAC___ ............................................(((((((((...........)))))))))................................... ( -8.52 = -8.21 + -0.31)

| Location | 12,000,431 – 12,000,521 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 56.85 |
| Shannon entropy | 0.90242 |
| G+C content | 0.49990 |
| Mean single sequence MFE | -22.36 |
| Consensus MFE | -8.52 |
| Energy contribution | -8.21 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 2.44 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.884527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
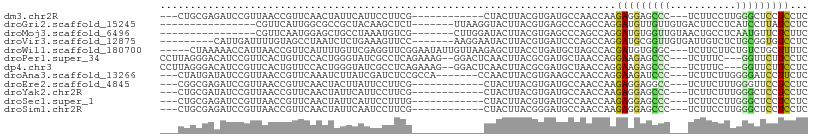
>dm3.chr2R 12000431 90 - 21146708 ---CUGCGAGAUCCGUUAACCGUUCAACUAUUCAUUCCUUCG------------CUACUUACGUGAUGCCAACCAAGAGGAGCCC---UCUUCCUUGGGCUCCUCCUC ---..(((.((...(((........)))...))......(((------------(.......))))))).......(((((((((---........)))))))))... ( -23.50, z-score = -2.47, R) >droGri2.scaffold_15245 16976952 85 - 18325388 ----------------CGUUCAUUGGCGCCGCUACAAGCUCU-------UUAAGGUACUUACGUGAGCCCAGCCAGGAUGUUGUUGUGACUUCCUCAUCCUUAUCCUC ----------------.......((((...((((((((..((-------....))..)))..)).)))...))))(((((.....((((.....))))...))))).. ( -15.90, z-score = 0.80, R) >droMoj3.scaffold_6496 16322874 85 + 26866924 ----------------CGUUCAAUGGAGCUGCCUAAAUGUCG-------CUUGGAUACUUACGUGAGCCCAGCCAGGAUGUGGUUGUAACUGCCUCAAUGUUCUCUUC ----------------(((((..(((.(((((.(((.((((.-------....)))).))).)).))))))....))))).(((.......))).............. ( -15.80, z-score = 1.55, R) >droVir3.scaffold_12875 17464457 92 - 20611582 ---------CAUUGAUUUUGUAGCCUAAUCUCUGAAAGUUCC-------AAGGAAUACUUACGUGAUCCCAGCCAGGAUGCGGUUGUGAUUGUCUCUGCGGUGUCCUC ---------....(((.((((((..(((((.......((((.-------...))))((...(((.((((......)))))))...)))))))...)))))).)))... ( -18.80, z-score = 0.63, R) >droWil1.scaffold_180700 4164296 100 - 6630534 -----CUAAAAACCAUUAACCGUUCAUUUUGUUCGAGGUUCGGAAUAUUGUUAAGAGCUUACCUGAUGCUAGCCACGAUGUGGGC---UCUUCUUCUGUCUGCUUUUC -----.............................((((..((((........((((((((((.(..((.....))..).))))))---))))......))))..)))) ( -15.64, z-score = 1.34, R) >droPer1.super_34 563932 100 - 916997 CCUUAGGGACAUCCGUUCACUGUUCCACUGGGUAUCGCCUCAGAAAG--GGACUCAACUUACGCGAUGCUAACCAGGAAGAGCCC---UCUUUC---GGUUCUUCCUC .....((..(((((((.....(((((.(((((.....)).)))...)--)))).......))).))))....))((((((((((.---......---)))))))))). ( -33.70, z-score = -2.27, R) >dp4.chr3 18617450 100 - 19779522 CCUUAGGGACAUCCGUUCACUGUUCCACUGGGUAUCGCCUCAGAAAG--GGACUCAACUUACGCGAUGCUAACCAGGAAGAGCCC---UCUUUC---GGUUCUUCCUC .....((..(((((((.....(((((.(((((.....)).)))...)--)))).......))).))))....))((((((((((.---......---)))))))))). ( -33.70, z-score = -2.27, R) >droAna3.scaffold_13266 4477216 95 + 19884421 ---CUAUGAUAUCCGUUAACCGUUCAAAUCUUAUCGAUCUCCGCCA-------CCAACUUACGUGAAGCCAACCAGGAAGAUCCC---UCUUCUUGGGGAUCCUUCUC ---....((((....................))))(((((..((((-------(........)))..))...(((((((((....---))))))))))))))...... ( -18.55, z-score = -0.53, R) >droEre2.scaffold_4845 8731945 90 + 22589142 ---CGGCGAGAUCCGUUAACCGUUCAACUACUUAUUCCUUCG------------CUACUUACGUGAUGCCAACCAAGAGGAGGCC---UCUUCUUUGGGUUCCUCCUC ---.((((.((..((.....)).))..............(((------------(.......))))))))......(((((((.(---((......)))..))))))) ( -21.60, z-score = -0.61, R) >droYak2.chr2R 4155905 90 + 21139217 ---CUGCGAUAUCCGUUAACCGUUCAACUAUUCAUUCCUUCG------------CUACUUACGUGAUGCCAACCAAGAGGAGCCC---UCUUCUUUGGGCUCCUCCUC ---..(((......(((........)))...........(((------------(.......))))))).......(((((((((---........)))))))))... ( -22.70, z-score = -2.91, R) >droSec1.super_1 9495943 90 - 14215200 ---CUGCGAGAUCCGUUAACCGUUCAACUAUUCAUUCCUUUG------------CUACUUACGUGAUGCCAACCAAGAGGAGCCC---UCUUCCUUGGGCUCCUCCUC ---....(((((((((.....((..................)------------).....))).))).........(((((((((---........)))))))))))) ( -23.17, z-score = -2.41, R) >droSim1.chr2R 10737936 90 - 19596830 ---CUGCGAGAUCCGUUAACCGUUCAACUAUUCAAUCCUUCG------------CUACUUACGGGAUGCCAACCAAGAGGAGCCC---UCUUCCUUGGGCUCCUCCUC ---....(((....((...((((...................------------......))))...)).......(((((((((---........)))))))))))) ( -25.21, z-score = -2.10, R) >consensus ___CUGCGAGAUCCGUUAACCGUUCAACUAUUUAUUCCUUCG____________CUACUUACGUGAUGCCAACCAGGAGGAGCCC___UCUUCCUUGGGCUCCUCCUC ............................................................................(((((((((...........)))))))))... ( -8.52 = -8.21 + -0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:26 2011