| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,998,121 – 11,998,182 |
| Length | 61 |
| Max. P | 0.530615 |

| Location | 11,998,121 – 11,998,182 |
|---|---|
| Length | 61 |
| Sequences | 12 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 88.83 |
| Shannon entropy | 0.22846 |
| G+C content | 0.53097 |
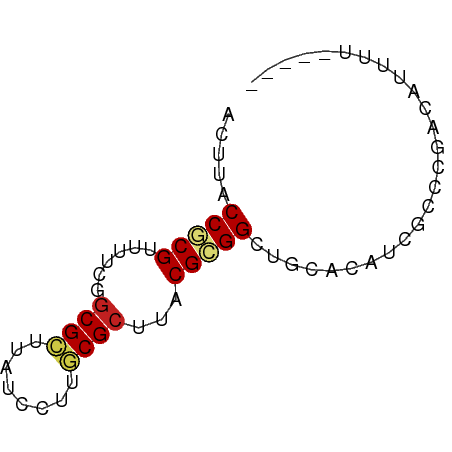
| Mean single sequence MFE | -14.95 |
| Consensus MFE | -11.44 |
| Energy contribution | -11.71 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.56 |
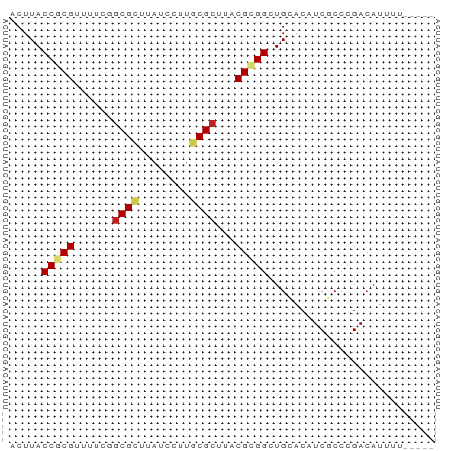
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.530615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
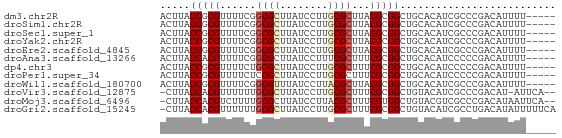
>dm3.chr2R 11998121 61 - 21146708 ACUUACCGCGUUUUCGGCGCUUAUCCUUGCGCUUACGCGGCUGCACAUCGCCCGACAUUUU----- .....((((((....(((((........))))).))))))..((.....))..........----- ( -18.40, z-score = -1.85, R) >droSim1.chr2R 10735553 61 - 19596830 ACUUACCGCGUUUUCGGCGCUUAUCCUUGCGCUUACGCGGCUGCACAUCGCCCGACAUUUU----- .....((((((....(((((........))))).))))))..((.....))..........----- ( -18.40, z-score = -1.85, R) >droSec1.super_1 9493617 61 - 14215200 ACUUACCGCGUUUUCGGCGCUUAUCCUUGCGCUUACGCGGCUGCACAUCGCCCGACAUUUU----- .....((((((....(((((........))))).))))))..((.....))..........----- ( -18.40, z-score = -1.85, R) >droYak2.chr2R 4153559 61 + 21139217 ACUUACCGCGUUUUCGGCGCUUAUCCUUGCGCUUACGCGGCUGCACAUCGCCCGACAUUUU----- .....((((((....(((((........))))).))))))..((.....))..........----- ( -18.40, z-score = -1.85, R) >droEre2.scaffold_4845 8729638 61 + 22589142 ACUUACCGCGUUUUCGGCGCUUAUCCUUGCGCUUACGCGGCUGCACAUCGCCCGACAUUUU----- .....((((((....(((((........))))).))))))..((.....))..........----- ( -18.40, z-score = -1.85, R) >droAna3.scaffold_13266 4474925 61 + 19884421 ACUUACCACGUUUUCGGCGCUUAUCCUUUCGCUUUCGCGGCUGCACAUCCCCCGACAUUUU----- .............((((........(..((((....))))..)........))))......----- ( -7.49, z-score = 0.59, R) >dp4.chr3 18615050 61 - 19779522 ACUUACCGCGUUUUCUGCGCUUAUCCUUGCGCUUUCGCGGCUGCACAUCCCCCGACAUUUU----- .....(((((......((((........))))...))))).....................----- ( -15.50, z-score = -2.65, R) >droPer1.super_34 561519 61 - 916997 ACUUACCGCGUUUUCUCCGCUUAUCCUUGCGCUUUCGCGGCUGCACAUCCCCCGACAUUUU----- .....(((((.......(((........)))....))))).....................----- ( -11.20, z-score = -1.43, R) >droWil1.scaffold_180700 4161390 61 - 6630534 ACUUACCGCGUUUUCGGCGUUUAUCCUUACGCUUACGCGGCUGCACAUCGCCCGACAUUUU----- .....((((((....(((((........))))).))))))..((.....))..........----- ( -16.00, z-score = -1.75, R) >droVir3.scaffold_12875 17460854 62 - 20611582 -CUUACCACGUUUUUUGCGCUUAUCCUUGCGCUUUCGCGGCUGUACAUCGCCCGACAU-AUUCA-- -...............((((........))))..(((.(((........))))))...-.....-- ( -13.80, z-score = -1.90, R) >droMoj3.scaffold_6496 16319839 63 + 26866924 -CUUACCACGUCUUUUGCGCUUAUCCUUACGCUUUCGUGGCUGUACGUCGCCCGACAUAAUUCA-- -....(((((......(((..........)))...)))))......(((....)))........-- ( -11.80, z-score = -1.27, R) >droGri2.scaffold_15245 16974454 65 - 18325388 -CUUACCACGUUUUUUGCGCUUAUCCUUGCGCUUUCGCGGCUGUACAUCGCCUGACAUAUUUUUCA -...............((((........))))..(((.(((........))))))........... ( -11.60, z-score = -1.08, R) >consensus ACUUACCGCGUUUUCGGCGCUUAUCCUUGCGCUUACGCGGCUGCACAUCGCCCGACAUUUU_____ .....(((((......((((........))))...))))).......................... (-11.44 = -11.71 + 0.26)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:24 2011