| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,986,346 – 11,986,414 |
| Length | 68 |
| Max. P | 0.995025 |

| Location | 11,986,346 – 11,986,414 |
|---|---|
| Length | 68 |
| Sequences | 6 |
| Columns | 75 |
| Reading direction | forward |
| Mean pairwise identity | 55.02 |
| Shannon entropy | 0.84405 |
| G+C content | 0.38152 |
| Mean single sequence MFE | -16.40 |
| Consensus MFE | -5.87 |
| Energy contribution | -7.68 |
| Covariance contribution | 1.82 |
| Combinations/Pair | 1.90 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.995025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
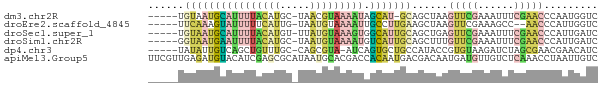
>dm3.chr2R 11986346 68 + 21146708 -----UGUAAUGCAUUUUACAUGC-UAACGUAAAAUAGCAU-GCAGCUAAGUUCGAAAUUUCGAACCCAAUGGUC -----((((.((((((((((....-....))))))).))))-)))((((.((((((....))))))....)))). ( -16.70, z-score = -2.57, R) >droEre2.scaffold_4845 8717836 67 - 22589142 -----UUCAAAGUAUUUUUCAUUG-UAAUGUAAAAUUGCCUUGAAGCUAAGUUCGAAAGCC--AACCCAUUGGUC -----(((((.(((.(((((((..-..))).)))).))).))))).............(((--((....))))). ( -10.20, z-score = -1.12, R) >droSec1.super_1 9482022 69 + 14215200 -----UGUAAUGCAUUUUACAUGU-UUAUGUAAAGUGGCAUUGCAGCUGAGUUCGAAAUUUCGAACCCAUUGAUC -----(((((((((((((((((..-..))))))))).))))))))..((.((((((....)))))).))...... ( -23.80, z-score = -4.62, R) >droSim1.chr2R 10721750 69 + 19596830 -----GGUAAUGAAUUUUACAUGC-UAAUGUAAAAUGUCAUUGCAGCUUUGUUCGAAAUUUCGAACCCAUUGAUC -----.((((((((((((((((..-..))))))))).)))))))......((((((....))))))......... ( -20.90, z-score = -4.57, R) >dp4.chr3 16418964 68 + 19779522 -----UAUAUUGUCAGCUGUUUGC-CAGCGUA-AUCAGUGCUGCCAUACCGUGUAAGAUCUAGCGAACGAACAUC -----....((((..(((((.((.-(((((..-.....))))).)).))(......)....)))..))))..... ( -11.10, z-score = 0.98, R) >apiMel3.Group5 290723 75 - 13386189 UUCGUUGAGAUGUACAUCGAGCGCAUAAUGCACGACCACAAUGACGACAAUGAUGUUGUCUCAAACCUAAUUGUC ....((((((((.((((((..(((((..((......))..))).))....))))))))))))))........... ( -15.70, z-score = -0.23, R) >consensus _____UGUAAUGUAUUUUACAUGC_UAAUGUAAAAUAGCAUUGCAGCUAAGUUCGAAAUCUCGAACCCAAUGAUC ......((((((((((((((((.....))))))))).)))))))......(((((......)))))......... ( -5.87 = -7.68 + 1.82)
| Location | 11,986,346 – 11,986,414 |
|---|---|
| Length | 68 |
| Sequences | 6 |
| Columns | 75 |
| Reading direction | reverse |
| Mean pairwise identity | 55.02 |
| Shannon entropy | 0.84405 |
| G+C content | 0.38152 |
| Mean single sequence MFE | -16.73 |
| Consensus MFE | -2.83 |
| Energy contribution | -6.25 |
| Covariance contribution | 3.42 |
| Combinations/Pair | 1.74 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.17 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.752588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
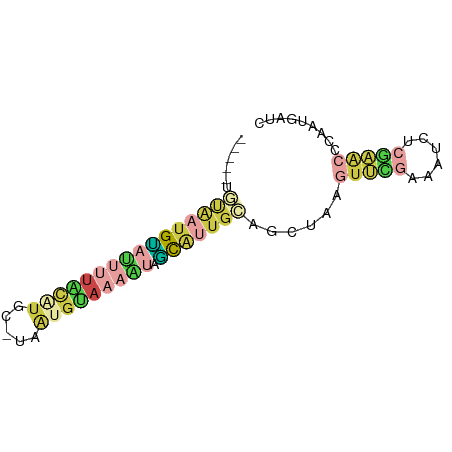
>dm3.chr2R 11986346 68 - 21146708 GACCAUUGGGUUCGAAAUUUCGAACUUAGCUGC-AUGCUAUUUUACGUUA-GCAUGUAAAAUGCAUUACA----- ......(((((((((....)))))))))(((((-((((((........))-)))))))....))......----- ( -21.40, z-score = -4.08, R) >droEre2.scaffold_4845 8717836 67 + 22589142 GACCAAUGGGUU--GGCUUUCGAACUUAGCUUCAAGGCAAUUUUACAUUA-CAAUGAAAAAUACUUUGAA----- ((((....))))--((((.........))))((((((..(((((.(((..-..))).))))).)))))).----- ( -10.90, z-score = -1.07, R) >droSec1.super_1 9482022 69 - 14215200 GAUCAAUGGGUUCGAAAUUUCGAACUCAGCUGCAAUGCCACUUUACAUAA-ACAUGUAAAAUGCAUUACA----- ......(((((((((....)))))))))...(.(((((...(((((((..-..)))))))..))))).).----- ( -19.60, z-score = -4.34, R) >droSim1.chr2R 10721750 69 - 19596830 GAUCAAUGGGUUCGAAAUUUCGAACAAAGCUGCAAUGACAUUUUACAUUA-GCAUGUAAAAUUCAUUACC----- .......((((((((....))))))........(((((.(((((((((..-..)))))))))))))).))----- ( -16.30, z-score = -3.26, R) >dp4.chr3 16418964 68 - 19779522 GAUGUUCGUUCGCUAGAUCUUACACGGUAUGGCAGCACUGAU-UACGCUG-GCAAACAGCUGACAAUAUA----- ..((((.(((.((((.(((......))).)))))))......-...((((-.....)))).)))).....----- ( -14.40, z-score = -0.02, R) >apiMel3.Group5 290723 75 + 13386189 GACAAUUAGGUUUGAGACAACAUCAUUGUCGUCAUUGUGGUCGUGCAUUAUGCGCUCGAUGUACAUCUCAACGAA ((((((...(((......)))...)))))).((.(((.(((.(((((((........)))))))))).))).)). ( -17.80, z-score = -0.09, R) >consensus GACCAAUGGGUUCGAAAUUUCGAACUUAGCUGCAAUGCCAUUUUACAUUA_GCAUGUAAAAUACAUUACA_____ ......(((((((((....))))))))).....(((((.(((((((((.....))))))))))))))........ ( -2.83 = -6.25 + 3.42)

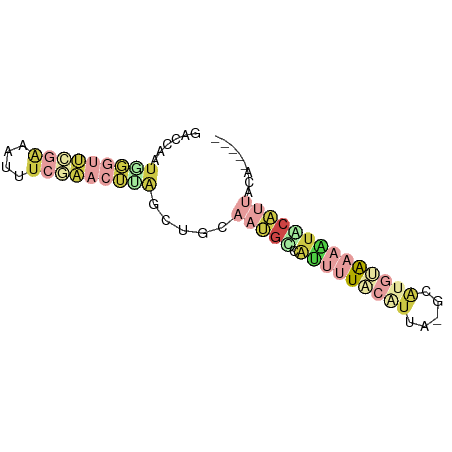
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:24 2011