
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,978,665 – 11,978,729 |
| Length | 64 |
| Max. P | 0.986967 |

| Location | 11,978,665 – 11,978,729 |
|---|---|
| Length | 64 |
| Sequences | 8 |
| Columns | 70 |
| Reading direction | forward |
| Mean pairwise identity | 68.85 |
| Shannon entropy | 0.58578 |
| G+C content | 0.54966 |
| Mean single sequence MFE | -16.78 |
| Consensus MFE | -8.49 |
| Energy contribution | -8.49 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
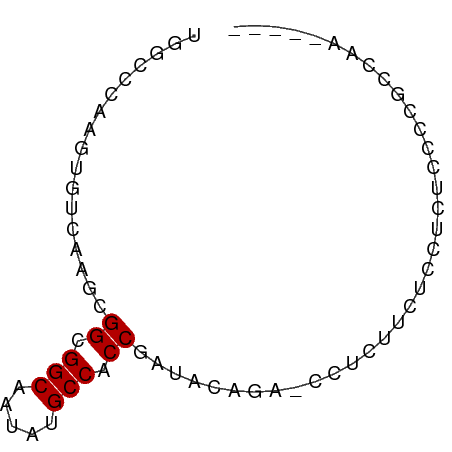
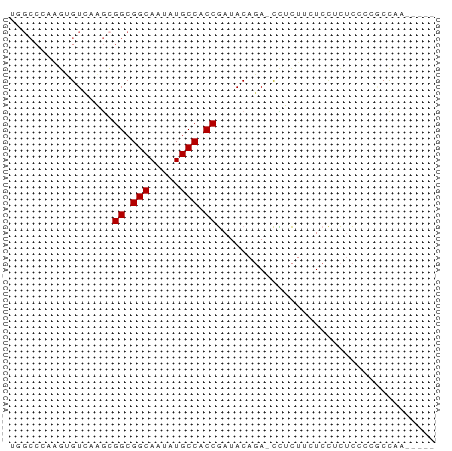
>dm3.chr2R 11978665 64 + 21146708 UGGCCCAAGUGUCAAGCGGCGGCAAUAUGCCACCGAUACAGA-CCUCUUGUCCUCUCCCUGCCAA----- ((((.((((.(((...(((.(((.....))).))).....))-)..))))..........)))).----- ( -18.80, z-score = -1.64, R) >droSim1.chr2R 10714193 64 + 19596830 UGGCCCAAGUGUCAAGCGGCGGCAAUGUGCCACCGAUACAGA-CCUCUUGUCCUCUCCCUGCCAA----- ((((.((((.(((...(((.(((.....))).))).....))-)..))))..........)))).----- ( -18.80, z-score = -1.16, R) >droSec1.super_1 9474482 64 + 14215200 UGGCCCAAGUGUCAAGCGGCGGCAAUAUGCCACCGAUACAGA-CCUCUUGUCCUCUCCCUGCCAA----- ((((.((((.(((...(((.(((.....))).))).....))-)..))))..........)))).----- ( -18.80, z-score = -1.64, R) >droYak2.chr2R 4133846 63 - 21139217 --GCCCAAGUGUCAAGCGGCGGCAAUAUGCCACCGAUACAGAACCUCUGUUCCUCUCCCAACCAA----- --......(((((....((.(((.....))).))))))).((((....)))).............----- ( -13.60, z-score = -1.46, R) >droEre2.scaffold_4845 8710003 64 - 22589142 UGGCCCAAGUGUCAAGCGGCGGCAAUAUGCCACCGAUACAGA-CCUCUGUUCCUCUCCCGGCCAA----- (((((...........(((.(((.....))).)))..((((.-...)))).........))))).----- ( -21.90, z-score = -2.19, R) >droAna3.scaffold_13266 4460056 64 - 19884421 UGGCCAAGCGGUGUCGGGGCGGCAAUAUGCCACCGAUACAGA-CUGCUCCUCCACUCCACACCAC----- (((...(((((((((((((((......)))).)))))....)-)))))...)))...........----- ( -21.90, z-score = -1.39, R) >dp4.chr3 16412226 62 + 19779522 -----AAAAAGUGUCA-GGCGGCAAUAUGCCACCAAUACAUAUUUGUCUCUCUCUC--GCUCUCUGUCUA -----.....((((..-((.(((.....))).)).)))).................--............ ( -10.20, z-score = -1.47, R) >droPer1.super_4 6812660 64 - 7162766 -----AAAAAGUGUCA-GGCGGCAAUAUGCCACCAAUACAUAUUUGUCUCUCUCUCUCGCUCUCUGUCUA -----.....((((..-((.(((.....))).)).))))............................... ( -10.20, z-score = -1.63, R) >consensus UGGCCCAAGUGUCAAGCGGCGGCAAUAUGCCACCGAUACAGA_CCUCUUCUCCUCUCCCCGCCAA_____ .................((.(((.....))).)).................................... ( -8.49 = -8.49 + -0.00)

| Location | 11,978,665 – 11,978,729 |
|---|---|
| Length | 64 |
| Sequences | 8 |
| Columns | 70 |
| Reading direction | reverse |
| Mean pairwise identity | 68.85 |
| Shannon entropy | 0.58578 |
| G+C content | 0.54966 |
| Mean single sequence MFE | -22.09 |
| Consensus MFE | -11.49 |
| Energy contribution | -11.49 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.986967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
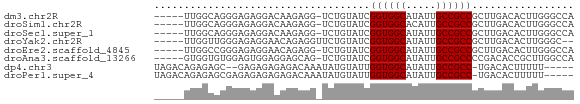
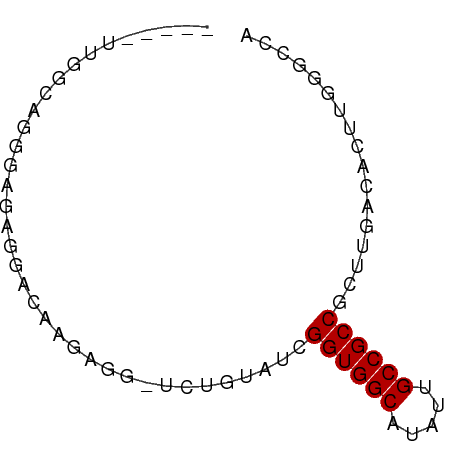
>dm3.chr2R 11978665 64 - 21146708 -----UUGGCAGGGAGAGGACAAGAGG-UCUGUAUCGGUGGCAUAUUGCCGCCGCUUGACACUUGGGCCA -----.((((...(((.((((.....)-)))((..(((((((.....)))))))....)).)))..)))) ( -23.80, z-score = -1.54, R) >droSim1.chr2R 10714193 64 - 19596830 -----UUGGCAGGGAGAGGACAAGAGG-UCUGUAUCGGUGGCACAUUGCCGCCGCUUGACACUUGGGCCA -----.((((...(((.((((.....)-)))((..(((((((.....)))))))....)).)))..)))) ( -23.80, z-score = -1.29, R) >droSec1.super_1 9474482 64 - 14215200 -----UUGGCAGGGAGAGGACAAGAGG-UCUGUAUCGGUGGCAUAUUGCCGCCGCUUGACACUUGGGCCA -----.((((...(((.((((.....)-)))((..(((((((.....)))))))....)).)))..)))) ( -23.80, z-score = -1.54, R) >droYak2.chr2R 4133846 63 + 21139217 -----UUGGUUGGGAGAGGAACAGAGGUUCUGUAUCGGUGGCAUAUUGCCGCCGCUUGACACUUGGGC-- -----...((..((...(((((....)))))....(((((((.....)))))))))..))........-- ( -22.50, z-score = -2.18, R) >droEre2.scaffold_4845 8710003 64 + 22589142 -----UUGGCCGGGAGAGGAACAGAGG-UCUGUAUCGGUGGCAUAUUGCCGCCGCUUGACACUUGGGCCA -----.(((((.(..(((..((((...-.))))..(((((((.....))))))))))..).....))))) ( -26.50, z-score = -2.07, R) >droAna3.scaffold_13266 4460056 64 + 19884421 -----GUGGUGUGGAGUGGAGGAGCAG-UCUGUAUCGGUGGCAUAUUGCCGCCCCGACACCGCUUGGCCA -----(((((((((.(((.(((.....-))).))).((((((.....)))))))).)))))))....... ( -25.50, z-score = -0.92, R) >dp4.chr3 16412226 62 - 19779522 UAGACAGAGAGC--GAGAGAGAGACAAAUAUGUAUUGGUGGCAUAUUGCCGCC-UGACACUUUUU----- .....(((((((--....)...........(((...((((((.....))))))-..)))))))))----- ( -15.40, z-score = -2.90, R) >droPer1.super_4 6812660 64 + 7162766 UAGACAGAGAGCGAGAGAGAGAGACAAAUAUGUAUUGGUGGCAUAUUGCCGCC-UGACACUUUUU----- .....(((((((....).............(((...((((((.....))))))-..)))))))))----- ( -15.40, z-score = -2.87, R) >consensus _____UUGGCAGGGAGAGGACAAGAGG_UCUGUAUCGGUGGCAUAUUGCCGCCGCUUGACACUUGGGCCA ....................................((((((.....))))))................. (-11.49 = -11.49 + -0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:21 2011