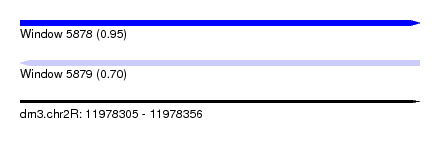
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,978,305 – 11,978,356 |
| Length | 51 |
| Max. P | 0.953278 |

| Location | 11,978,305 – 11,978,356 |
|---|---|
| Length | 51 |
| Sequences | 5 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 76.12 |
| Shannon entropy | 0.40736 |
| G+C content | 0.37043 |
| Mean single sequence MFE | -14.86 |
| Consensus MFE | -8.78 |
| Energy contribution | -8.98 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.953278 |
| Prediction | RNA |
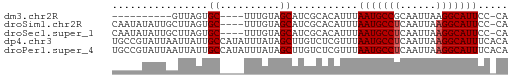
Download alignment: ClustalW | MAF
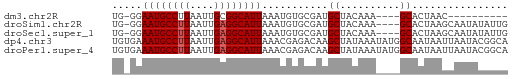
>dm3.chr2R 11978305 51 + 21146708 UG-GGAAUGCCUUAAUUGCGGCAUUAAAUGUGCGAUGCUACAAA----GCACUAAC---------- ..-..((((((........))))))....((((...........----))))....---------- ( -11.70, z-score = -1.12, R) >droSim1.chr2R 10713864 61 + 19596830 UG-GGAAUGCCUUAAUUGAGGCAUUAAAUGUGCGAUGCUACAAA----GCACUAAGCAAUAUAUUG ..-..((((((((....)))))))).....(((..((((....)----)))....)))........ ( -15.60, z-score = -2.02, R) >droSec1.super_1 9474147 61 + 14215200 UG-GGAAUGCCUUAAUUGAGGCAUUAAAUGUGCGAUGCUACAAA----GCACUAAGCAAUAUAUUG ..-..((((((((....)))))))).....(((..((((....)----)))....)))........ ( -15.60, z-score = -2.02, R) >dp4.chr3 16411820 66 + 19779522 UGUGAAAUGCCUUAAUUGAGGCAUUAAACGAGACAAGCUAUAAAUAUGGCAAUAAUUAAUACGGCA .((..((((((((....))))))))..)).......(((((....)))))................ ( -15.70, z-score = -2.89, R) >droPer1.super_4 6812254 66 - 7162766 UGUGAAAUGCCUUAAUUGAGGCAUUAAACGAGACAAGCUAUAAAUAUGGCAAUAAUUAAUACGGCA .((..((((((((....))))))))..)).......(((((....)))))................ ( -15.70, z-score = -2.89, R) >consensus UG_GGAAUGCCUUAAUUGAGGCAUUAAAUGUGCGAUGCUACAAA____GCACUAAGCAAUACAGCA .....((((((((....))))))))......................................... ( -8.78 = -8.98 + 0.20)
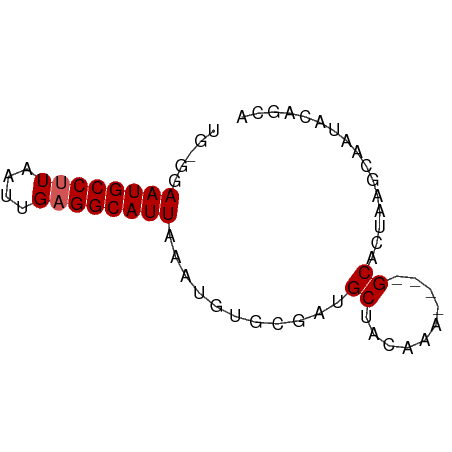
| Location | 11,978,305 – 11,978,356 |
|---|---|
| Length | 51 |
| Sequences | 5 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 76.12 |
| Shannon entropy | 0.40736 |
| G+C content | 0.37043 |
| Mean single sequence MFE | -12.56 |
| Consensus MFE | -6.84 |
| Energy contribution | -7.04 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.695826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
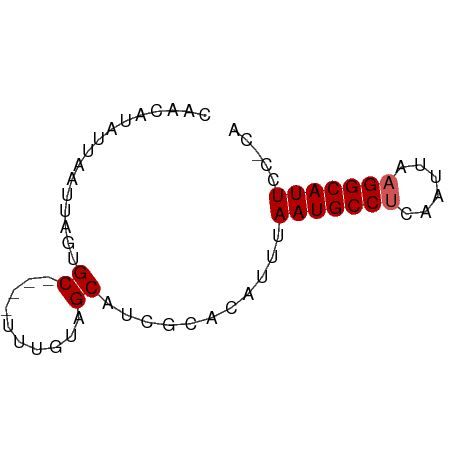
>dm3.chr2R 11978305 51 - 21146708 ----------GUUAGUGC----UUUGUAGCAUCGCACAUUUAAUGCCGCAAUUAAGGCAUUCC-CA ----------....((((----..((...))..))))....((((((........))))))..-.. ( -12.20, z-score = -1.63, R) >droSim1.chr2R 10713864 61 - 19596830 CAAUAUAUUGCUUAGUGC----UUUGUAGCAUCGCACAUUUAAUGCCUCAAUUAAGGCAUUCC-CA ........(((...((((----(....))))).))).....(((((((......)))))))..-.. ( -14.00, z-score = -2.15, R) >droSec1.super_1 9474147 61 - 14215200 CAAUAUAUUGCUUAGUGC----UUUGUAGCAUCGCACAUUUAAUGCCUCAAUUAAGGCAUUCC-CA ........(((...((((----(....))))).))).....(((((((......)))))))..-.. ( -14.00, z-score = -2.15, R) >dp4.chr3 16411820 66 - 19779522 UGCCGUAUUAAUUAUUGCCAUAUUUAUAGCUUGUCUCGUUUAAUGCCUCAAUUAAGGCAUUUCACA .((.(((.....))).))...................((..(((((((......)))))))..)). ( -11.30, z-score = -1.48, R) >droPer1.super_4 6812254 66 + 7162766 UGCCGUAUUAAUUAUUGCCAUAUUUAUAGCUUGUCUCGUUUAAUGCCUCAAUUAAGGCAUUUCACA .((.(((.....))).))...................((..(((((((......)))))))..)). ( -11.30, z-score = -1.48, R) >consensus CAACAUAUUAAUUAGUGC____UUUGUAGCAUCGCACAUUUAAUGCCUCAAUUAAGGCAUUCC_CA .........................................(((((((......)))))))..... ( -6.84 = -7.04 + 0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:19 2011