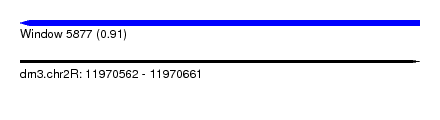
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,970,562 – 11,970,661 |
| Length | 99 |
| Max. P | 0.910370 |

| Location | 11,970,562 – 11,970,661 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 75.55 |
| Shannon entropy | 0.50044 |
| G+C content | 0.34306 |
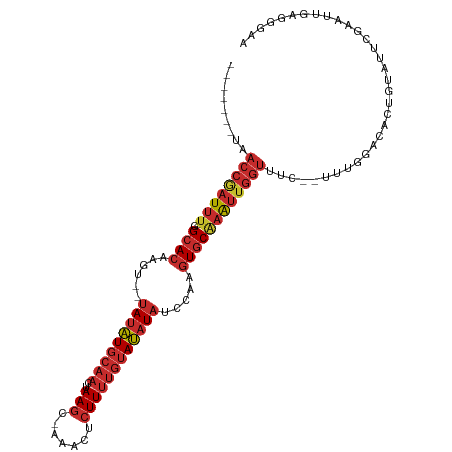
| Mean single sequence MFE | -21.09 |
| Consensus MFE | -12.55 |
| Energy contribution | -13.80 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.910370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11970562 99 - 21146708 -------UAACCGAUUU-GGCACAAGU--UAUAUGCAACUAAGC-AAACUCUUUUGUAUAUAUCCAAGUGCAAAUUGGUUUC--UUUGGACACUGUAUUCGAAUUGAGGGAA -------.(((((((((-(.(((....--(((((((((..(((.-.....)))))))))))).....)))))))))))))((--(((.((..(.......)..)).))))). ( -21.90, z-score = -1.29, R) >droWil1.scaffold_181141 1401885 93 - 5303230 --------------UCUUGGCACCAUCUUUAACUACAACUAAUA-UAAUUUUUACAAAGCUCUGCUAUUUCUAGCUGUUU----UCUAUACAAUAAAUUAAAAAGGAAUGGA --------------........((((((((.........(((..-......))).(((((...((((....)))).))))----).................)))).)))). ( -9.00, z-score = 0.37, R) >droPer1.super_4 6803676 111 + 7162766 UAUACAUCUACCGAUUU-GGCACAACUUAUAUAUGCAACUAAGCAAAACUCUUUUGUAUAUAUCCAAGUGCGAAUUGGUUUCUUUUUGGACUAGCUAUUCAAAUGGAAGCAA ...........((((((-(.(((.....((((((((((...((.....))...))))))))))....))))))))))((((((.((((((.......)))))).)))))).. ( -25.30, z-score = -2.64, R) >dp4.chr3 16403248 111 - 19779522 UAUACAUCUACCGAUUU-GGCACAACUUAUAUAUGCAACUAAGCAAAACUCUUUUGUAUAUAUCCAAGUGCGAAUUGGUUUCUUUUUGGACUAGCUAUUCAAAUGGAAGCAA ...........((((((-(.(((.....((((((((((...((.....))...))))))))))....))))))))))((((((.((((((.......)))))).)))))).. ( -25.30, z-score = -2.64, R) >droAna3.scaffold_13266 4452853 86 + 19884421 -------UAACCGAUUUCGGCACAAGU--UAUAUGCAACUAAGC-CAAACCUUUUGUAUAUAUUCGAGUGCAAAUUGGUUUC--UUUCGAGUGGGUCA-------------- -------.((((((((...((((..(.--(((((((((..(((.-.....))))))))))))..)..)))).))))))))..--..............-------------- ( -19.80, z-score = -1.45, R) >droEre2.scaffold_4845 8701869 99 + 22589142 -------UAACCAAUUU-GGCACAAGU--UAUAUGCAACUAAGC-AAACUCUUUUGUAUAUAUCCAAGUGCAAAUUGGUUUC--UUUGGACACUGUAUUCGAAUUGAGGGAA -------.(((((((((-(.(((....--(((((((((..(((.-.....)))))))))))).....)))))))))))))((--(((.((..(.......)..)).))))). ( -22.20, z-score = -1.48, R) >droYak2.chr2R 4125572 99 + 21139217 -------UAACCGAUUU-GGCACAAGU--UAUAUGCAACUAAGC-AAACUCUUUUGUAUAUAUCCAAGUGCAAAUUGGUUUC--UUUGGACACUGUAUUCGAAUUGAGGGAA -------.(((((((((-(.(((....--(((((((((..(((.-.....)))))))))))).....)))))))))))))((--(((.((..(.......)..)).))))). ( -21.90, z-score = -1.29, R) >droSec1.super_1 9466425 99 - 14215200 -------UAACCGAUUU-GGCACAAGU--UAUAUGCAACUAAGC-AAACUCUUUUGUAUAUAUCCAAGUGCAAAUUGGUUUC--UUUGGACACCGUAUUCGAAUUGAGGGAA -------.(((((((((-(.(((....--(((((((((..(((.-.....)))))))))))).....)))))))))))))((--(((.((...((....))..)).))))). ( -23.60, z-score = -1.80, R) >droSim1.chr2R_random 2492326 99 - 2996586 -------UAACCGAUUU-GGCACAAGU--UAUAUGCAACUAAGC-AAACUCUUUUGUAUAUAUCCAAGUGCAAAUUGGUUUC--UUUGUACACUGUAUUCGAAUUGAGGGAA -------..((((((((-(.(((....--(((((((((..(((.-.....)))))))))))).....))))))))))))(((--((((................))))))). ( -20.79, z-score = -1.03, R) >consensus _______UAACCGAUUU_GGCACAAGU__UAUAUGCAACUAAGC_AAACUCUUUUGUAUAUAUCCAAGUGCAAAUUGGUUUC__UUUGGACACUGUAUUCGAAUUGAGGGAA .........((((((((..((((......(((((((((..(((.......)))))))))))).....))))))))))))................................. (-12.55 = -13.80 + 1.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:18 2011