
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,962,572 – 11,962,666 |
| Length | 94 |
| Max. P | 0.859792 |

| Location | 11,962,572 – 11,962,666 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 68.74 |
| Shannon entropy | 0.54966 |
| G+C content | 0.50391 |
| Mean single sequence MFE | -28.42 |
| Consensus MFE | -15.22 |
| Energy contribution | -16.70 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
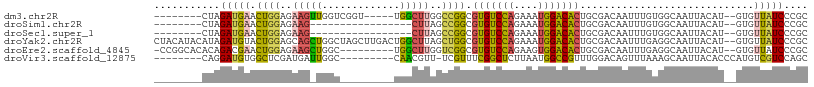
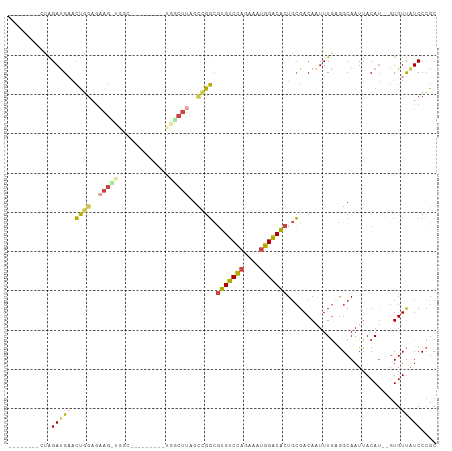
>dm3.chr2R 11962572 94 + 21146708 --------CUAGAUGAACUGGAGAAGUUGGUCGGU-----UGGCUUGGCCGGCGUGUCCAGAAAUGGACACUGCGACAAUUUGUGGCAAUUACAU--GUGUUAUCCCGC --------...(((.((((.....)))).))).((-----((((...))))))(((((((....)))))))(((.((.....)).))).......--............ ( -32.80, z-score = -2.12, R) >droSim1.chr2R 10700440 82 + 19596830 --------CUAGAUGAACUGGAGAAG-----------------CUUAGCCGGCGUGUCCAGAAAUGGACACUGCGACAAUUUGUGGCAAUUACAU--GUGUUAUCCCGC --------...(((((((((.....(-----------------((.....)))(((((((....)))))))(((.((.....)).)))....)).--)).))))).... ( -23.80, z-score = -1.61, R) >droSec1.super_1 9458495 82 + 14215200 --------CUAGAUGAACUGGAGAAG-----------------CUUAGCCGGCGUGUCCAGAAAUGGACACUGCGACAAUUUGUGGCAAUUACAU--GUGUUAUCCCGC --------...(((((((((.....(-----------------((.....)))(((((((....)))))))(((.((.....)).)))....)).--)).))))).... ( -23.80, z-score = -1.61, R) >droYak2.chr2R 4116788 107 - 21139217 CUACAUACAUAGAUGUACUGGAGCAGCUGGCUAGCUUGACUGGCUUAGCUGGCGUGUCCAGAAAUGGACACUGCGACAAUUUGAGGCAAUUACAU--GUGUUAUCCCGC .(((((......)))))..((((((((..(((((((.....))).))))..))(((((((....)))))))))).......(((.(((......)--)).))))))... ( -38.80, z-score = -2.95, R) >droEre2.scaffold_4845 8694039 97 - 22589142 -CCGGCACACAGACGAACUGGAGAAGCUGGC---------UGGCUUGGUCGGCGUGUCCAGAAGUGGACACUGCGACAAUUUGAGGCAAUUACAU--GUGUUAUCCCGC -.(((.(((((..((((.((..(..((((((---------(.....)))))))(((((((....)))))))..)..)).)))).(.......).)--))))....))). ( -29.80, z-score = -0.09, R) >droVir3.scaffold_12875 17418683 91 + 20611582 --------CAGGAUGUGGCUCGAUGAUUGGC---------CAACGUU-UCGUUUCGGCUCUUAAUGGCCGUUUGGACAGUUUAAAGCAAUUACACCCAUGUCGUCCAGC --------(..(((((((((........)))---------).)))))-..)...(((((......))))).((((((.((.....))....(((....))).)))))). ( -21.50, z-score = 0.53, R) >consensus ________CUAGAUGAACUGGAGAAG_UGGC_________UGGCUUAGCCGGCGUGUCCAGAAAUGGACACUGCGACAAUUUGAGGCAAUUACAU__GUGUUAUCCCGC ...........(((((.((((..((((((...........))))))..)))).(((((((....))))))).............................))))).... (-15.22 = -16.70 + 1.48)

| Location | 11,962,572 – 11,962,666 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 68.74 |
| Shannon entropy | 0.54966 |
| G+C content | 0.50391 |
| Mean single sequence MFE | -22.80 |
| Consensus MFE | -13.91 |
| Energy contribution | -14.05 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.28 |
| Mean z-score | -0.67 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.586105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
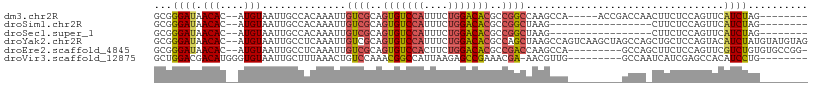
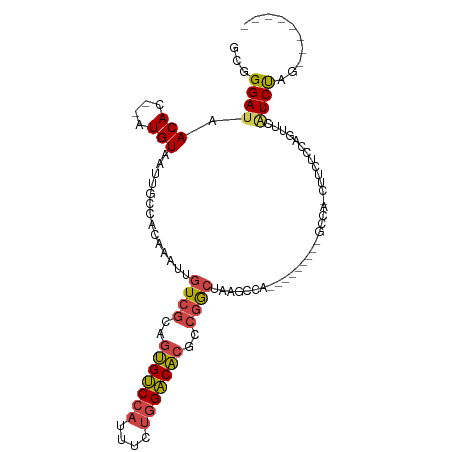
>dm3.chr2R 11962572 94 - 21146708 GCGGGAUAACAC--AUGUAAUUGCCACAAAUUGUCGCAGUGUCCAUUUCUGGACACGCCGGCCAAGCCA-----ACCGACCAACUUCUCCAGUUCAUCUAG-------- ...(((((((..--.(((.......)))....(((((.(((((((....))))))))).(((...))).-----...)))...........))).))))..-------- ( -21.30, z-score = -0.83, R) >droSim1.chr2R 10700440 82 - 19596830 GCGGGAUAACAC--AUGUAAUUGCCACAAAUUGUCGCAGUGUCCAUUUCUGGACACGCCGGCUAAG-----------------CUUCUCCAGUUCAUCUAG-------- ((.((.......--.......(((.((.....)).)))(((((((....))))))).)).))....-----------------..................-------- ( -20.20, z-score = -1.20, R) >droSec1.super_1 9458495 82 - 14215200 GCGGGAUAACAC--AUGUAAUUGCCACAAAUUGUCGCAGUGUCCAUUUCUGGACACGCCGGCUAAG-----------------CUUCUCCAGUUCAUCUAG-------- ((.((.......--.......(((.((.....)).)))(((((((....))))))).)).))....-----------------..................-------- ( -20.20, z-score = -1.20, R) >droYak2.chr2R 4116788 107 + 21139217 GCGGGAUAACAC--AUGUAAUUGCCUCAAAUUGUCGCAGUGUCCAUUUCUGGACACGCCAGCUAAGCCAGUCAAGCUAGCCAGCUGCUCCAGUACAUCUAUGUAUGUAG ...(((......--.......(((..(.....)..)))(((((((....)))))))((.((((..((.((.....)).)).))))))))).(((((....))))).... ( -26.70, z-score = -0.14, R) >droEre2.scaffold_4845 8694039 97 + 22589142 GCGGGAUAACAC--AUGUAAUUGCCUCAAAUUGUCGCAGUGUCCACUUCUGGACACGCCGACCAAGCCA---------GCCAGCUUCUCCAGUUCGUCUGUGUGCCGG- .(((.(((.((.--(((.(((((.........((((..(((((((....)))))))..)))).((((..---------....))))...)))))))).))))).))).- ( -29.40, z-score = -1.20, R) >droVir3.scaffold_12875 17418683 91 - 20611582 GCUGGACGACAUGGGUGUAAUUGCUUUAAACUGUCCAAACGGCCAUUAAGAGCCGAAACGA-AACGUUG---------GCCAAUCAUCGAGCCACAUCCUG-------- ..((((((....((((......)))).....))))))..((((........))))......-...((.(---------(((.......).)))))......-------- ( -19.00, z-score = 0.52, R) >consensus GCGGGAUAACAC__AUGUAAUUGCCACAAAUUGUCGCAGUGUCCAUUUCUGGACACGCCGGCUAAGCCA_________GCCA_CUUCUCCAGUUCAUCUAG________ ...((((.(((....)))..............((((..(((((((....)))))))..)))).................................)))).......... (-13.91 = -14.05 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:17 2011