
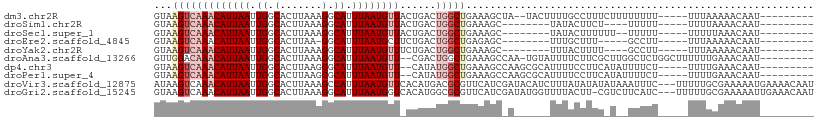
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,955,998 – 11,956,092 |
| Length | 94 |
| Max. P | 0.532179 |

| Location | 11,955,998 – 11,956,092 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 70.38 |
| Shannon entropy | 0.58235 |
| G+C content | 0.31608 |
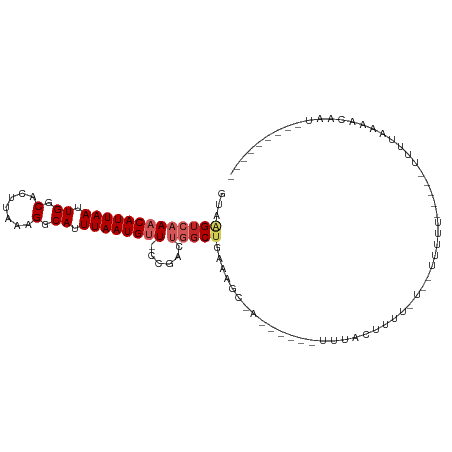
| Mean single sequence MFE | -17.54 |
| Consensus MFE | -7.77 |
| Energy contribution | -8.60 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.532179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11955998 94 + 21146708 GUAAGUCAAACAUUAAUUGGCACUUAAAGGCAUUUAAUGUUACUGACUGGCUGAAAGCUA--UACUUUUGCCUUUCUUUUUUUU-----UUUAAAAACAAU--------- ...(((((((((((((.((.(.......).)).))))))))..)))))(((.(((((...--..))))))))............-----............--------- ( -19.70, z-score = -2.20, R) >droSim1.chr2R 10691450 84 + 19596830 GUAAGUCAAACAUUAAUUGGCACUUAAAGGCAUUUAAUGUUACUGACUGGCUGAAAGC--------UAUACUUCU----UUUUU-----UUUUAAAACAAU--------- ((((((((((((((((.((.(.......).)).))))))))..)))))(((.....))--------).)))....----.....-----............--------- ( -16.20, z-score = -1.96, R) >droSec1.super_1 9452023 86 + 14215200 GUAAGUCAAACAUUAAUUGGCACUUAAAGGCAUUUAAUGUUACUGACUGGCUGAAAGC--------UAUACUUUUUU--UUUUU-----UUUUUAAACAAU--------- ((((((((((((((((.((.(.......).)).))))))))..)))))(((.....))--------).)))......--.....-----............--------- ( -16.20, z-score = -1.91, R) >droEre2.scaffold_4845 8687142 82 - 22589142 GUAAGUCAAACAUUAAUUGGCACUUAA-GGCAUUUAAUGCUUCUGACUGGCUGAGAGC--------UUUGCUUU-----GCCUU-----UUUAAAAACAAU--------- ...(((((((((((((.((.(......-).)).)))))).)).)))))(((..(((((--------...)))))-----)))..-----............--------- ( -17.40, z-score = -0.66, R) >droYak2.chr2R 4110135 84 - 21139217 GUAAGUCAAACAUUAAUUGGCACUUAAAGGCAUUUAAUGUUUCUGACUGGCUGAAAGC--------UUUACUUUU----GCCUU-----UUUAAAAACAAU--------- ...(((((((((((((.((.(.......).)).))))))))..)))))(((.(((((.--------....)))))----)))..-----............--------- ( -19.70, z-score = -2.54, R) >droAna3.scaffold_13266 4439966 98 - 19884421 GUUGGACAAACAUUAAUUGGCACUUAAAGGCAUUUAAUGUU--CGACUGGCUGAAAGCCAA-UGUAUUUUCUUCGCUUGGCUCUGGCUUUUUUGAAACAAU--------- ((..(.(.((((((((.((.(.......).)).))))))))--.).)..)).((((((((.-.((..............))..))))))))..........--------- ( -22.34, z-score = -1.24, R) >dp4.chr3 16389507 94 + 19779522 GUAAGUCAAACAUUAAUUGGCACUUAAGGGCAUUUAAUGUU--CAUAUGGCUGAAAGCCAAGCGCAUUUUCCUUCAUAUUUUCU-----UUUUGAAACAAU--------- ((...(((((((((((.((.(.(....)).)).))))))).--....((((.....))))........................-----..)))).))...--------- ( -14.70, z-score = 0.17, R) >droPer1.super_4 6789668 94 - 7162766 GUAAGUCAAACAUUAAUUGGCACUUAAGGGCAUUUAAUGUU--CAUAUGGCUGAAAGCCAAGCGCAUUUUCCUUCAUAUUUUCU-----UUUUGAAACAAU--------- ((...(((((((((((.((.(.(....)).)).))))))).--....((((.....))))........................-----..)))).))...--------- ( -14.70, z-score = 0.17, R) >droVir3.scaffold_12875 17400790 107 + 20611582 AUAAGUCAAACAUUAAUUGGCACUUAAAGCCAUUUAAUGUUCACAUGACGCGUUCAUCGAUACAUCUUUAUAUAUAUAAAUUUC---UUUUUGCGAAAAAUGAAAACAAU ....((((((((((((.((((.......)))).))))))))....))))(..(((((.........(((((....)))))((((---.......)))).)))))..)... ( -19.40, z-score = -2.45, R) >droGri2.scaffold_15245 16922955 106 + 18325388 GUAAGUCAAACAUUAAUUGGCACUUAAAGGCAUUUAAUGGUCACAUGGCGCGUUCAUCGAUAUGGUUUUACUU-CGUCUUCAUC---UUUUUGCGAAAAAUUGAAACAAU ....(((...((((((.((.(.......).)).))))))(((....))).........)))...((((((.((-(((.......---.....)))))....))))))... ( -15.10, z-score = 0.77, R) >consensus GUAAGUCAAACAUUAAUUGGCACUUAAAGGCAUUUAAUGUU_CCGACUGGCUGAAAGC_A______UUUACUUUU_U__UUUUU_____UUUUAAAACAAU_________ ...(((((((((((((.((.(.......).)).))))))))......))))).......................................................... ( -7.77 = -8.60 + 0.83)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:15 2011