| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,952,737 – 11,952,844 |
| Length | 107 |
| Max. P | 0.999242 |

| Location | 11,952,737 – 11,952,844 |
|---|---|
| Length | 107 |
| Sequences | 12 |
| Columns | 130 |
| Reading direction | forward |
| Mean pairwise identity | 74.97 |
| Shannon entropy | 0.50885 |
| G+C content | 0.40113 |
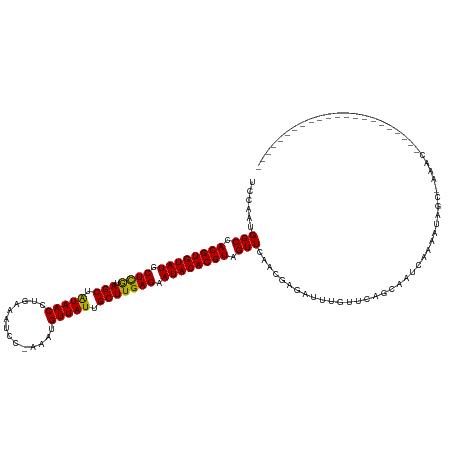
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -21.39 |
| Energy contribution | -21.56 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.73 |
| SVM RNA-class probability | 0.999242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
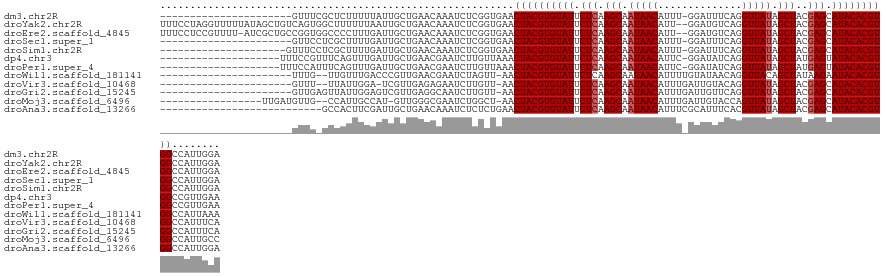
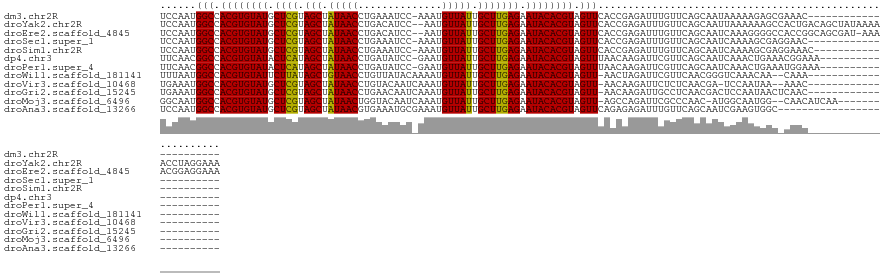
>dm3.chr2R 11952737 107 + 21146708 ----------------------GUUUCGCUCUUUUUAUUGCUGAACAAAUCUCGGUGAACUACGUGUAUUCUCAAGCAAUAACAUUU-GGAUUUCAGGUUAUAGCUACGAGCAUACACGUGGCCAUUGGA ----------------------((((.((..........)).)))).....((((((..((((((((((.(((.(((.(((((...(-(.....)).))))).)))..))).)))))))))).)))))). ( -31.20, z-score = -3.25, R) >droYak2.chr2R 4106864 128 - 21139217 UUUCCUAGGUUUUUAUAGCUGUCAGUGGCUUUUUUAAUUGCUGAACAAAUCUCGGUGAACUACGUGUAUUCUCAAGCAAUAACAUU--GGAUGUCAGGUUAUAGCUACGAGCAUACACGUGGCCAUUGGA .......((((.....)))).((((((((........(..((((.......))))..)..(((((((((.(((.(((.(((((.((--(.....)))))))).)))..))).))))))))))))))))). ( -39.20, z-score = -3.07, R) >droEre2.scaffold_4845 8683948 127 - 22589142 UUUCCUCCGUUUU-AUCGCUGCCGGUGGCCCCUUUGAUUGCUGAACAAAUCUCGGUGAACUACGUGUAUUCUCAAGCAAUAACAUU--GGAUGUCAGGUUAUAGCUACGAGCAUACACGUGGCCAUUGGA .............-.......(((((((((.......(..((((.......))))..)...((((((((.(((.(((.(((((.((--(.....)))))))).)))..))).))))))))))))))))). ( -42.00, z-score = -2.86, R) >droSec1.super_1 9448865 107 + 14215200 ----------------------GUUCCUCGCUUUUGAUUGCUGAACAAAUCUCGGUGAACUACGUGUAUUCUCAAGCAAUAACAUUU-GGAUUUCAGGUUAUAGCUACGAGCAUACACGUGGCCAUUGGA ----------------------..((((((((...((((........))))..))))).((((((((((.(((.(((.(((((...(-(.....)).))))).)))..))).)))))))))).....))) ( -31.90, z-score = -2.83, R) >droSim1.chr2R 10688331 108 + 19596830 ---------------------GUUUCCUCGCUUUUGAUUGCUGAACAAAUCUCGGUGAACUACGUGUAUUCUCAAGCAAUAACAUUU-GGAUUUCAGGUUAUAGCUACGAGCAUACACGUGGCCAUUGGA ---------------------...((((((((...((((........))))..))))).((((((((((.(((.(((.(((((...(-(.....)).))))).)))..))).)))))))))).....))) ( -31.90, z-score = -2.83, R) >dp4.chr3 16385880 109 + 19779522 --------------------UUUCCGUUUCAGUUUGAUUGCUGAACGAAUCUUGUUAAACUACGUGUAUUCUCAAGCAAUAACAUUC-GGAUAUCAGGUUAUAGCUAUGAGUAUACACGUGGCCGUUGAA --------------------....((((.((((......)))))))).......((((.((((((((((.(((((((.(((((....-.(....)..))))).))).)))).))))))))))...)))). ( -33.10, z-score = -3.58, R) >droPer1.super_4 6785678 109 - 7162766 --------------------UUUCCAUUUCAGUUUGAUUGCUGAACGAAUCUUGUUAAACUACGUGUAUUCUCAAGCAAUAACAUUC-GGAUAUCAGGUUAUAGCUAUGAGUAUACACGUGGCCGUUGAA --------------------.......((((((......)))))).........((((.((((((((((.(((((((.(((((....-.(....)..))))).))).)))).))))))))))...)))). ( -31.10, z-score = -3.22, R) >droWil1.scaffold_181141 1110988 105 + 5303230 ----------------------UUUG--UUGUUUGACCCGUUGAACGAAUCUAGUU-AACUACGUGUAUUCUCAAGCAAUAACAUUUUGUAUAACAGGUUACAGCUAUAAGAAUACACGUGGCCAUUAAA ----------------------....--(((((..(....)..)))))........-..(((((((((((((..(((..((((....((.....)).))))..)))...)))))))))))))........ ( -27.90, z-score = -3.13, R) >droVir3.scaffold_10468 1946 104 + 3271 ----------------------GUUU--UUAUUGGA-UCGUUGAGAGAAUCUUGUU-AACUACGUGUAUUCUCAAGCAAUAACAUUUGAUUGUACAGGUUAUAGCUACGAGCAUACACGUGGCCAUUUCA ----------------------....--....(((.-.....(((.....)))...-..((((((((((.(((.(((.(((((...((......)).))))).)))..))).)))))))))))))..... ( -26.00, z-score = -1.98, R) >droGri2.scaffold_15245 16918474 107 + 18325388 ----------------------GUUGAGUUAUUGGAGUCGUUGAGGCAAUCUUGUU-AACUACGUGUAUUCUCAAGCAAUAACAUUUGAUUGUUCAGGUUAUAGCUACGAGCAUACACGUGGCCAUUUCA ----------------------..((((....(((.(((.....))).........-..((((((((((.(((.(((.(((((...(((....))).))))).)))..))).))))))))))))).)))) ( -31.60, z-score = -2.81, R) >droMoj3.scaffold_6496 16256750 109 - 26866924 -----------------UUGAUGUUG--CCAUUGCCAU-GUUGGGCGAAUCUGGCU-AACUACGUGUAUUCUCAAGCAAUAACAUUUGAUUGUACCAGUUAUAGCUACGAGCAUACACGUGGCCAUUGCC -----------------..((((..(--((((((((..-....)))))...)))).-..((((((((((.(((.(((.(((((..............))))).)))..))).)))))))))).))))... ( -34.84, z-score = -2.38, R) >droAna3.scaffold_13266 4437075 103 - 19884421 ---------------------------GCCACUUCGAUUGCUGAACAAAUCUCUCUGAACUACGUGUAUUCUCAAGCAAUAACAUUUCGCAUUUCACGUUAUAGCUACGAGCAUACACGUGGCCAUUGGA ---------------------------.(((....((((........))))........((((((((((.(((.(((.(((((..............))))).)))..))).))))))))))....))). ( -26.44, z-score = -2.75, R) >consensus ______________________GUUG_GCCAUUUUGAUUGCUGAACAAAUCUCGGUGAACUACGUGUAUUCUCAAGCAAUAACAUUU_GGAUUUCAGGUUAUAGCUACGAGCAUACACGUGGCCAUUGGA ...........................................................((((((((((.(((.(((.(((((..............))))).)))..))).))))))))))........ (-21.39 = -21.56 + 0.17)
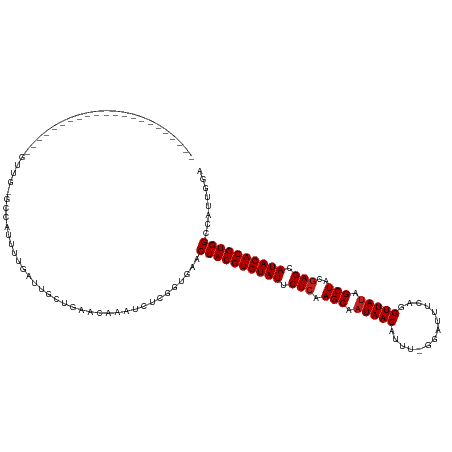
| Location | 11,952,737 – 11,952,844 |
|---|---|
| Length | 107 |
| Sequences | 12 |
| Columns | 130 |
| Reading direction | reverse |
| Mean pairwise identity | 74.97 |
| Shannon entropy | 0.50885 |
| G+C content | 0.40113 |
| Mean single sequence MFE | -29.75 |
| Consensus MFE | -21.70 |
| Energy contribution | -21.37 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.67 |
| SVM RNA-class probability | 0.999148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11952737 107 - 21146708 UCCAAUGGCCACGUGUAUGCUCGUAGCUAUAACCUGAAAUCC-AAAUGUUAUUGCUUGAGAAUACACGUAGUUCACCGAGAUUUGUUCAGCAAUAAAAAGAGCGAAAC---------------------- .....((..(((((((((.((((.(((.(((((.((.....)-)...))))).))))))).)))))))).)..))......(((((((...........)))))))..---------------------- ( -27.10, z-score = -2.79, R) >droYak2.chr2R 4106864 128 + 21139217 UCCAAUGGCCACGUGUAUGCUCGUAGCUAUAACCUGACAUCC--AAUGUUAUUGCUUGAGAAUACACGUAGUUCACCGAGAUUUGUUCAGCAAUUAAAAAAGCCACUGACAGCUAUAAAAACCUAGGAAA (((..((..(((((((((.((((.(((.(((((.((.....)--)..))))).))))))).)))))))).)..)).......((((.(((...............))))))).............))).. ( -28.26, z-score = -1.58, R) >droEre2.scaffold_4845 8683948 127 + 22589142 UCCAAUGGCCACGUGUAUGCUCGUAGCUAUAACCUGACAUCC--AAUGUUAUUGCUUGAGAAUACACGUAGUUCACCGAGAUUUGUUCAGCAAUCAAAGGGGCCACCGGCAGCGAU-AAAACGGAGGAAA (((..(((((((((((((.((((.(((.(((((.((.....)--)..))))).))))))).))))))))......((.....(((.....))).....)))))))(((........-....))).))).. ( -40.30, z-score = -2.63, R) >droSec1.super_1 9448865 107 - 14215200 UCCAAUGGCCACGUGUAUGCUCGUAGCUAUAACCUGAAAUCC-AAAUGUUAUUGCUUGAGAAUACACGUAGUUCACCGAGAUUUGUUCAGCAAUCAAAAGCGAGGAAC---------------------- (((..((..(((((((((.((((.(((.(((((.((.....)-)...))))).))))))).)))))))).)..))....((((.(.....)))))........)))..---------------------- ( -27.00, z-score = -2.00, R) >droSim1.chr2R 10688331 108 - 19596830 UCCAAUGGCCACGUGUAUGCUCGUAGCUAUAACCUGAAAUCC-AAAUGUUAUUGCUUGAGAAUACACGUAGUUCACCGAGAUUUGUUCAGCAAUCAAAAGCGAGGAAAC--------------------- (((..((..(((((((((.((((.(((.(((((.((.....)-)...))))).))))))).)))))))).)..))....((((.(.....)))))........)))...--------------------- ( -27.00, z-score = -2.03, R) >dp4.chr3 16385880 109 - 19779522 UUCAACGGCCACGUGUAUACUCAUAGCUAUAACCUGAUAUCC-GAAUGUUAUUGCUUGAGAAUACACGUAGUUUAACAAGAUUCGUUCAGCAAUCAAACUGAAACGGAAA-------------------- ......(((.((((((((.((((.(((.(((((.........-....))))).))))))).)))))))).)))........(((((((((........))).))))))..-------------------- ( -29.02, z-score = -3.41, R) >droPer1.super_4 6785678 109 + 7162766 UUCAACGGCCACGUGUAUACUCAUAGCUAUAACCUGAUAUCC-GAAUGUUAUUGCUUGAGAAUACACGUAGUUUAACAAGAUUCGUUCAGCAAUCAAACUGAAAUGGAAA-------------------- ......(((.((((((((.((((.(((.(((((.........-....))))).))))))).)))))))).)))........(((((((((........))).))))))..-------------------- ( -27.22, z-score = -2.81, R) >droWil1.scaffold_181141 1110988 105 - 5303230 UUUAAUGGCCACGUGUAUUCUUAUAGCUGUAACCUGUUAUACAAAAUGUUAUUGCUUGAGAAUACACGUAGUU-AACUAGAUUCGUUCAACGGGUCAAACAA--CAAA---------------------- .....((((.(((((((((((((.(((.(((((.((.....))....))))).)))))))))))))))).)))-)....((((((.....))))))......--....---------------------- ( -32.80, z-score = -4.73, R) >droVir3.scaffold_10468 1946 104 - 3271 UGAAAUGGCCACGUGUAUGCUCGUAGCUAUAACCUGUACAAUCAAAUGUUAUUGCUUGAGAAUACACGUAGUU-AACAAGAUUCUCUCAACGA-UCCAAUAA--AAAC---------------------- .....((((.((((((((.((((.(((.(((((.((......))...))))).))))))).)))))))).)))-)....((((........))-))......--....---------------------- ( -24.30, z-score = -2.80, R) >droGri2.scaffold_15245 16918474 107 - 18325388 UGAAAUGGCCACGUGUAUGCUCGUAGCUAUAACCUGAACAAUCAAAUGUUAUUGCUUGAGAAUACACGUAGUU-AACAAGAUUGCCUCAACGACUCCAAUAACUCAAC---------------------- .....((((.((((((((.((((.(((.(((((.(((....)))...))))).))))))).)))))))).)))-).................................---------------------- ( -25.00, z-score = -2.67, R) >droMoj3.scaffold_6496 16256750 109 + 26866924 GGCAAUGGCCACGUGUAUGCUCGUAGCUAUAACUGGUACAAUCAAAUGUUAUUGCUUGAGAAUACACGUAGUU-AGCCAGAUUCGCCCAAC-AUGGCAAUGG--CAACAUCAA----------------- (((....)))((((((((.((((.(((.(((((..............))))).))))))).)))))))).(((-.((((.....(((....-..)))..)))--)))).....----------------- ( -36.44, z-score = -2.99, R) >droAna3.scaffold_13266 4437075 103 + 19884421 UCCAAUGGCCACGUGUAUGCUCGUAGCUAUAACGUGAAAUGCGAAAUGUUAUUGCUUGAGAAUACACGUAGUUCAGAGAGAUUUGUUCAGCAAUCGAAGUGGC--------------------------- .......(((((((((((.((((.(((.(((((((..........))))))).))))))).))))))((.(..((((....))))..).)).......)))))--------------------------- ( -32.50, z-score = -2.85, R) >consensus UCCAAUGGCCACGUGUAUGCUCGUAGCUAUAACCUGAAAUCC_AAAUGUUAUUGCUUGAGAAUACACGUAGUUCAACGAGAUUUGUUCAGCAAUCAAAAUAGC_AAAC______________________ ......(((.((((((((.((((.(((.(((((..............))))).))))))).)))))))).)))......................................................... (-21.70 = -21.37 + -0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:14 2011