
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,939,420 – 11,939,518 |
| Length | 98 |
| Max. P | 0.792908 |

| Location | 11,939,420 – 11,939,518 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 72.77 |
| Shannon entropy | 0.53651 |
| G+C content | 0.54331 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -19.82 |
| Energy contribution | -20.49 |
| Covariance contribution | 0.66 |
| Combinations/Pair | 1.47 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.792908 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 11939420 98 + 21146708 AUGGGGCUUACUGGCCCGAUCC---UUCUGCUCCUCCUGCUUCCGUAUUUGCUCGCCUGUUUUUGCGGUUAACUUGGUCUGCGAAAGGCAGGCA----GUCAAGU ...(((((....))))).....---....((.......))......(((((((.((((((((((((((..........)))))))).)))))))----).))))) ( -31.00, z-score = -0.58, R) >droSim1.chr2R 10677249 98 + 19596830 GUGGGGCUUACUGGCCCGGUCC---UUCUGCUCCUCCUGCUUCCGUAUUUGCUCGCCUGUUUUUGUGGUUAACUUGGUCUGCGAAAGGCAGGCA----GUCAAGU ...(((((....)))))((...---....((.......))..))..(((((((.(((((((((((..(..........)..))))).)))))))----).))))) ( -29.10, z-score = 0.26, R) >droSec1.super_1 9438446 98 + 14215200 GUGGGGCUUACUGGCCCGAUCC---UUCUGCUCCUCUUGCUUCCGUAUUUGCUCGCCUGUUUUUGCGGUUAACUUGGUCUGCGAAAGGCAGGCA----GUCAAGU ((.(((((....))))).))..---....((.......))......(((((((.((((((((((((((..........)))))))).)))))))----).))))) ( -31.30, z-score = -0.70, R) >droYak2.chr2R 4096167 99 - 21139217 GUGGGGCUUACUGGCCCGAUCC---UUCUGCUCC---UGCUUCCGUAUUUGCUCGCCUGUUUUUGCGGUUAACUUGGUCUGCAAAAGGCAGGCAGGCAGUCAAGU ...(((((....)))))(((..---....((...---.)).........(((..((((((((((((((..........)))))))).))))))..)))))).... ( -35.10, z-score = -1.02, R) >droEre2.scaffold_4845 8673458 96 - 22589142 GUGGGGCUUACUGGCCCGAUCC---UUCUGCUCCC--UGCUUCCGUAUUUGCUCGCCUGUUUUUGCGGUUAACUUGGUCUGCGAAAGGCAGGCA----GUCAAGU ((.(((((....))))).))..---....((....--.))......(((((((.((((((((((((((..........)))))))).)))))))----).))))) ( -31.60, z-score = -0.66, R) >droAna3.scaffold_13266 4428334 87 - 19884421 UUCCAGCUCCUGCUCCCGCUCCAGCUCCAGACCCUUCUGCUUCCGUAUUUGCUCGCCUGUUUUUGCGGUUAACUUGGUCCGUAGAAG------------------ ....((((...((....))...)))).......(((((((..(((..((.((.(((........))))).))..)))...)))))))------------------ ( -19.90, z-score = -1.41, R) >droVir3.scaffold_12875 17382365 94 + 20611582 ACUGGUCGUACACUCCCGACUA---GAGCACUCUUACUUAUUCCUUUCGUGCU-GCCUGCUCCUGCGGUUA-UGGGUUACGCAAGGGACCUGCUUUGAA------ .(((((((........))))))---)(((((.................)))))-((((((....))))...-.((((..(....)..))))))......------ ( -27.43, z-score = -1.33, R) >consensus GUGGGGCUUACUGGCCCGAUCC___UUCUGCUCCUCCUGCUUCCGUAUUUGCUCGCCUGUUUUUGCGGUUAACUUGGUCUGCGAAAGGCAGGCA____GUCAAGU ...(((((....))))).....................................((((((((((((((..........))))))))).)))))............ (-19.82 = -20.49 + 0.66)


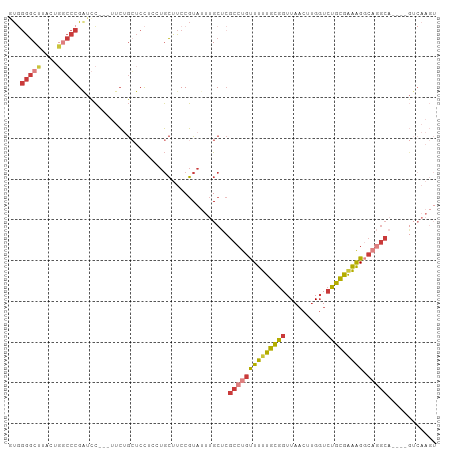
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:11 2011