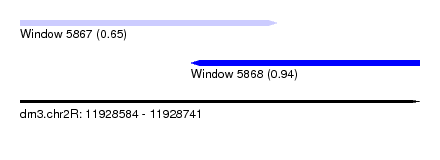
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,928,584 – 11,928,741 |
| Length | 157 |
| Max. P | 0.944867 |

| Location | 11,928,584 – 11,928,685 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 82.92 |
| Shannon entropy | 0.29109 |
| G+C content | 0.55753 |
| Mean single sequence MFE | -28.42 |
| Consensus MFE | -22.70 |
| Energy contribution | -22.22 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.645332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
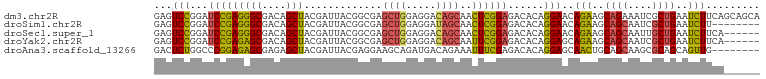
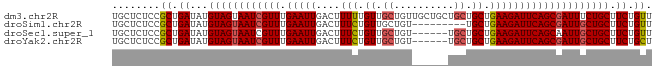
>dm3.chr2R 11928584 101 + 21146708 GAGUCCGGAUCCGAGGGCGACAGCUACGAUUACGGCGAGCUGGAGGACAGCAACUCGGAGACACAGGAACAGAAGCAGAAAUCGCUGAAUCUUCAGCAGCA ...((((..(((((((((....))).............((((.....))))..))))))..)...)))......((.(....)(((((....))))).)). ( -30.60, z-score = -1.36, R) >droSim1.chr2R 10667209 93 + 19596830 GAGUCCGGAUCCGAGGGCGACAGCUACGAUUACGGCGAGCUGGAGGAUAGCAACUCGGAGACACAGGAACAGAAGCAGCAAUCGCUGAAUCUU-------- ...((((..((((((.((..(((((.((....))...))))).......))..))))))..)...)))..(((..((((....))))..))).-------- ( -29.30, z-score = -1.82, R) >droSec1.super_1 9427546 95 + 14215200 GAGUCCGGAUCCGAGGGCGACAGCUACGAUUACGGCGAGCUGGAGGACAGCAACUCGGAGACACAGGAACAGAAGCAGCAAUUGCUGAAUCUUCA------ ...((((..(((((((((....))).............((((.....))))..))))))..)...)))...((((((((....))))...)))).------ ( -30.80, z-score = -2.13, R) >droYak2.chr2R 4085181 95 - 21139217 GAGUCCGGAUCCGAGAGCGACAGCUACGAUUACGGCGAGCUGGAGGACAGCAAUUCGGAGACACAGGAGCAGAAGCAGCAAUCGCUGAAUCUUCA------ ...((((..(((((((((....))).............((((.....))))..))))))..)...)))(.(((..((((....))))..))).).------ ( -28.30, z-score = -0.93, R) >droAna3.scaffold_13266 4417079 93 - 19884421 GACUCUGGCCCGGAGAGCGAGAGCUACGAUUACGAGGAAGCAGAUGACAGAAAUUUCGAGACACAGGAGCAACUGCAGCAAGCGCAGCAGUUG-------- ...((((.((((..((((....))).).....)).))...)))).........((((........))))(((((((.((....)).)))))))-------- ( -23.10, z-score = -0.33, R) >consensus GAGUCCGGAUCCGAGGGCGACAGCUACGAUUACGGCGAGCUGGAGGACAGCAACUCGGAGACACAGGAACAGAAGCAGCAAUCGCUGAAUCUUCA______ ...(((...(((((((((....))).............((((.....))))..))))))......)))..(((..((((....))))..)))......... (-22.70 = -22.22 + -0.48)
| Location | 11,928,651 – 11,928,741 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 92.53 |
| Shannon entropy | 0.11718 |
| G+C content | 0.42478 |
| Mean single sequence MFE | -22.32 |
| Consensus MFE | -19.98 |
| Energy contribution | -20.60 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
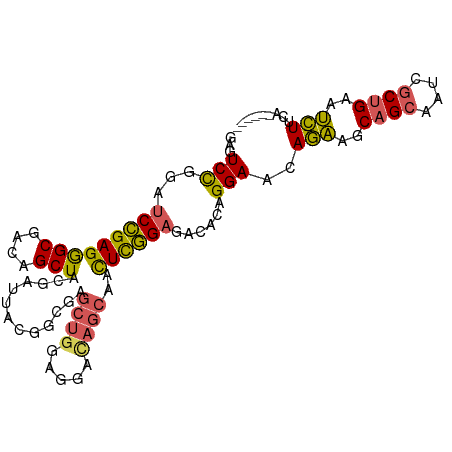
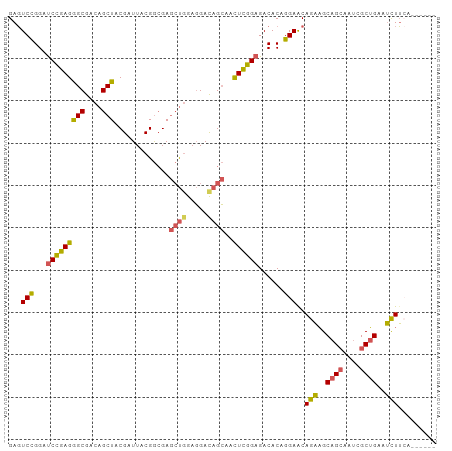
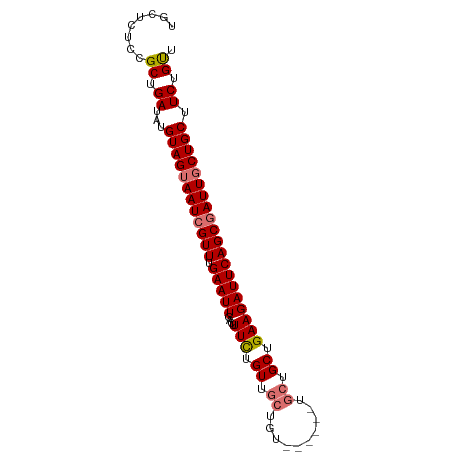
>dm3.chr2R 11928651 90 - 21146708 UGCUCUCCGCUGAUAUGUAGUAAUCGUUUGAAUUGACUUUUUGUUGCUGUUGCUGCUGCUGCUGAAGAUUCAGCGAUUUCUGCUUCUGUU ........((.((...((((.(((((((.((((...((((..((.((.((....)).)).)).))))))))))))))).)))).)).)). ( -20.20, z-score = -0.93, R) >droSim1.chr2R 10667276 81 - 19596830 UGCUCUCCGCUGAUAUGUAGUAAUCGUUUGAAUUGACUUUCUGUUGCUGU---------UGCUGAAGAUUCAGCGAUUGCUGCUUCUGUU ........((.((...((((((((((((.((((((((.....)))((...---------.))....))))))))))))))))).)).)). ( -22.60, z-score = -2.85, R) >droSec1.super_1 9427613 84 - 14215200 UGCUCUCCGCUGAUAUGUAGUAAUCGUUUGAAUUGACUUUCUGUUGCUGU------UGCUGCUGAAGAUUCAGCAAUUGCUGCUUCUGUU .((.....))......((((((((.((..(((......)))....)).))------))))))...(((..((((....))))..)))... ( -19.10, z-score = -1.09, R) >droYak2.chr2R 4085248 84 + 21139217 UGCUCUCCGCUGAUAUGUAGUAAUCGUUUGAAUUGACUUUCUGUUGCUGU------UGCUGCUGAAGAUUCAGCGAUUGCUGCUUCUGCU ........((.((...((((((((((((.(((((....(((.((.((...------.)).)).)))))))))))))))))))).)).)). ( -27.40, z-score = -3.97, R) >consensus UGCUCUCCGCUGAUAUGUAGUAAUCGUUUGAAUUGACUUUCUGUUGCUGU______UGCUGCUGAAGAUUCAGCGAUUGCUGCUUCUGUU ........((.((...((((((((((((.(((((....(((.((.((..........)).)).)))))))))))))))))))).)).)). (-19.98 = -20.60 + 0.62)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:10 2011