| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,925,394 – 11,925,444 |
| Length | 50 |
| Max. P | 0.592643 |

| Location | 11,925,394 – 11,925,444 |
|---|---|
| Length | 50 |
| Sequences | 10 |
| Columns | 51 |
| Reading direction | reverse |
| Mean pairwise identity | 96.46 |
| Shannon entropy | 0.07782 |
| G+C content | 0.34337 |
| Mean single sequence MFE | -8.71 |
| Consensus MFE | -6.92 |
| Energy contribution | -6.76 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.592643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
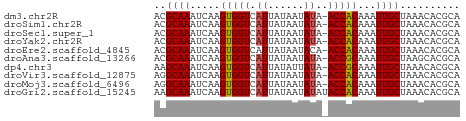
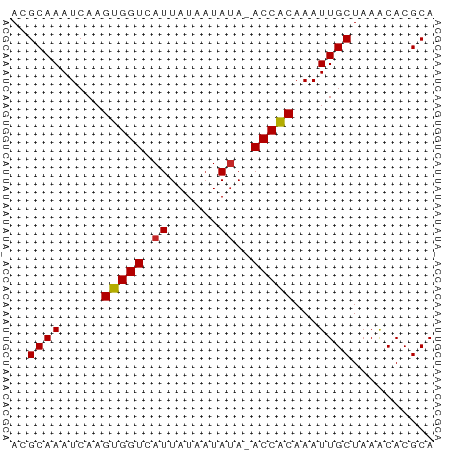
>dm3.chr2R 11925394 50 - 21146708 ACGCAAAUCAAGUGGUCAUUAUAAUAUA-ACCACAAAUUGCUAAACACGCA ..((((.....(((((.((......)).-)))))...)))).......... ( -8.50, z-score = -2.29, R) >droSim1.chr2R 10664032 50 - 19596830 ACGCAAAUCAAGUGGUCAUUAUAAUAUA-ACCACAAAUUGCUAAACACGCA ..((((.....(((((.((......)).-)))))...)))).......... ( -8.50, z-score = -2.29, R) >droSec1.super_1 9424382 50 - 14215200 ACGCAAAUCAAGUGGUCAUUAUAAUAUA-ACCACAAAUUGCUAAACACGCA ..((((.....(((((.((......)).-)))))...)))).......... ( -8.50, z-score = -2.29, R) >droYak2.chr2R 4082007 50 + 21139217 ACGCAAAUCAAGUGGUCAUUAUAAUAUA-ACCACAAAUUGCUAAACACGCA ..((((.....(((((.((......)).-)))))...)))).......... ( -8.50, z-score = -2.29, R) >droEre2.scaffold_4845 8656411 50 + 22589142 ACGCAAAUCAAGUGGUCAUUAUAAUACA-ACCACAAAUUGCUAAACACGCA ..((((.....(((((............-)))))...)))).......... ( -7.20, z-score = -1.60, R) >droAna3.scaffold_13266 4414559 50 + 19884421 ACGCAAAUCAAGUGGUCAUUAUAAUAUA-ACCGCAAAUUGCUAAGCACGCA ..((.......(((((.((......)).-)))))....(((...))).)). ( -8.90, z-score = -1.12, R) >dp4.chr3 16359205 50 - 19779522 AAGCAAAUCAAGUGGUCAUUAUAUUAUA-ACCGCAAAUUGCUAAACACGCA .(((((.....(((((.((......)).-)))))...)))))......... ( -9.20, z-score = -1.91, R) >droVir3.scaffold_12875 17365825 50 - 20611582 AGGCAAAUCAAGUGGUCAUUAUAAUAUA-ACCACAAAUUGCUAAACACGCA .(((((.....(((((.((......)).-)))))...)))))......... ( -9.60, z-score = -2.54, R) >droMoj3.scaffold_6496 16222767 50 + 26866924 AGGCAAAUCAAGUGGUCAUUAUAAUAUA-ACCACAAAUUGCUAAACACGCA .(((((.....(((((.((......)).-)))))...)))))......... ( -9.60, z-score = -2.54, R) >droGri2.scaffold_15245 16885670 51 - 18325388 AAGCAAAUCAAGUGGUCAUUAUAAUAUAUACCACAAAUUGCUAAACACGCA .(((((.....(((((.((........)))))))...)))))......... ( -8.60, z-score = -2.37, R) >consensus ACGCAAAUCAAGUGGUCAUUAUAAUAUA_ACCACAAAUUGCUAAACACGCA ..((((.....(((((.............)))))...)))).......... ( -6.92 = -6.76 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:08 2011