| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,821,499 – 11,821,599 |
| Length | 100 |
| Max. P | 0.923361 |

| Location | 11,821,499 – 11,821,599 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.11 |
| Shannon entropy | 0.60942 |
| G+C content | 0.33036 |
| Mean single sequence MFE | -22.93 |
| Consensus MFE | -6.70 |
| Energy contribution | -7.03 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
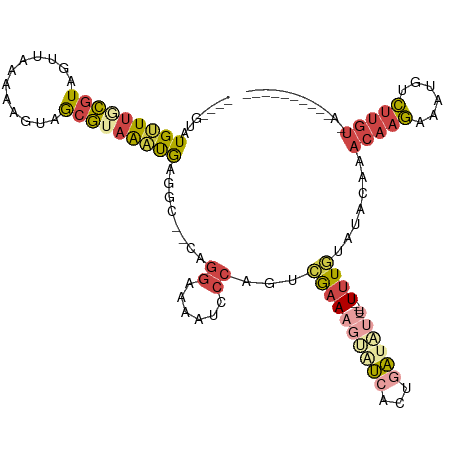
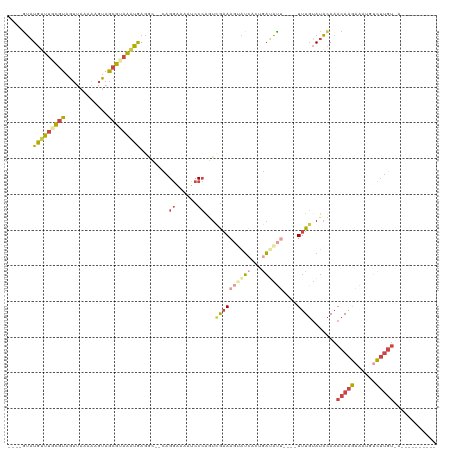
>dm3.chr2R 11821499 100 - 21146708 ----GUAUGAUUGCGUAGUUAAAAAGUAGCGUAAAUGAGGC--CAGGAAAAUCCCGGUCGAAAGUAUCAAUGAUAUU----UUUGUAUACAAACAAGAAAUGUCUUGUUA---------- ----(((((.((((((..((....))..))))))....(((--(.((.....)).))))(((((((((...))))))----))).))))).(((((((....))))))).---------- ( -26.00, z-score = -3.92, R) >droWil1.scaffold_180745 1437907 94 - 2843958 ----GGAUGAUCUUCUAUAGAAGUCGAAAUAUAGUCGAUAC--CCGAAAAAUCCAAAUGGCAAGGGGAUCAGUUCUU----UUUUAAUUUACACAGAAAUUCAU---------------- ----..((((..((((....((((..(((...((..(((.(--((.......((....))...))).)))..))...----)))..))))....))))..))))---------------- ( -15.00, z-score = 0.26, R) >droSim1.chr2R 10560134 100 - 19596830 ----GUAUGUUUGCGUAGUUAAAA-GUAGCGUAAAUGACGC--CAGGAAAAUCCCGGUCAAAAGUAUCACUGAUAUU----UUUGUAUACAAACAAGAAAUUUCUUGUUGU--------- ----((((((((((((........-...))))))))...((--(.((.....)).)))((((((((((...))))))----)))).)))).(((((((....)))))))..--------- ( -25.10, z-score = -3.27, R) >droSec1.super_1 9321716 102 - 14215200 GUACAUAUGUUUGCGUAGUUAAAAAGUAGCGUAAAUGAGGC--CAGGGAAAUCCCGGUCGAAAGUAUCACUGAUAUU----UUUGUAUACAAACAAGAAAUUUCUUGU------------ ((((....((((((((..((....))..))))))))..(((--(.(((....)))))))(((((((((...))))))----)))))))....((((((....))))))------------ ( -27.80, z-score = -3.55, R) >droYak2.chr2R 3978543 111 + 21139217 ----GUUUGUUUUCGUAGUUAAAAUGUAGCGUAAAUAAGCG--CAGGAAAAUCCCAUUCGAAAUAAUCUCGGAAACCU---UUUGUAUACAAACAAGAAAAGUCUUGUGAAAUAUGUAAG ----....(((((((..((((.......((((......)))--).((......))........))))..)))))))((---(.(((((....((((((....))))))...))))).))) ( -22.00, z-score = -2.28, R) >droEre2.scaffold_4845 8552845 116 + 22589142 ----GUAUGUUUACGUAGUUAAAAUGUAGCGUAAAUAAGAAAUAAGGGAAAACCCAGUAGAAAUAAUCUCUAAAACGUUUGUUUGUAUACAAACAAGAAAUGACUUGUGAAAUAUGUAAG ----((((.((((((.(((((.......((((.............(((....)))..((((.......))))..))))(((((((....)))))))....)))))))))))))))..... ( -21.70, z-score = -2.40, R) >consensus ____GUAUGUUUGCGUAGUUAAAAAGUAGCGUAAAUGAGGC__CAGGAAAAUCCCAGUCGAAAGUAUCACUGAUAUU____UUUGUAUACAAACAAGAAAUGUCUUGU_A__________ .......(((((((((............)))))))))........((......)).....................................(((((......)))))............ ( -6.70 = -7.03 + 0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:26:05 2011