
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,816,382 – 11,816,449 |
| Length | 67 |
| Max. P | 0.843618 |

| Location | 11,816,382 – 11,816,449 |
|---|---|
| Length | 67 |
| Sequences | 11 |
| Columns | 80 |
| Reading direction | reverse |
| Mean pairwise identity | 76.08 |
| Shannon entropy | 0.46616 |
| G+C content | 0.39268 |
| Mean single sequence MFE | -12.95 |
| Consensus MFE | -9.10 |
| Energy contribution | -9.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.843618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11816382 67 - 21146708 -----UCGUGCCUUAAAAAAUUAUUAACGGAAGUACAUUCACCUGAUCAGUUGAAUGAACAUGAGCUCUGCU-------- -----(((((................((....)).((((((.((....)).))))))..)))))........-------- ( -13.00, z-score = -1.55, R) >droSim1.chr2R 10555069 67 - 19596830 -----UCGUGCCUUAAAAAAUUAUUAACGGAAGUACAUUCACCUGAUCAGUUGAAUGAACAUGAGCUCUGCU-------- -----(((((................((....)).((((((.((....)).))))))..)))))........-------- ( -13.00, z-score = -1.55, R) >droSec1.super_1 9316639 67 - 14215200 -----UCGUGCCUUAAAAAAUUAUUAACGGAAGUACAUUCACCUGAUCAGUUGAAUGAACAUGAGCUCUGCU-------- -----(((((................((....)).((((((.((....)).))))))..)))))........-------- ( -13.00, z-score = -1.55, R) >droYak2.chr2R 3973239 67 + 21139217 -----UCGUGCCUUAACAAAUUAUUAACGGAAGUACAUUCACCUGAUCAGUUGAAUGAACAUGAGCUCUGCU-------- -----(((((................((....)).((((((.((....)).))))))..)))))........-------- ( -13.00, z-score = -1.30, R) >droEre2.scaffold_4845 8547465 67 + 22589142 -----UCGUGCCUUAACAAAUUAUUAACGGAAGUACAUUCACCUGAUCAGUUGAAUGAACAUGAGCUCUGCU-------- -----(((((................((....)).((((((.((....)).))))))..)))))........-------- ( -13.00, z-score = -1.30, R) >dp4.chr3 17874765 80 - 19779522 UCAUGGAUGAAAAAAAAGAGUUAUUAACGGAAGUACAUUCACCUGAUCAGUUGAAUGAACAUGCGCCUCGCCACACUGUU ...(((.(((................((....)).((((((.((....)).))))))..........))))))....... ( -13.10, z-score = 0.17, R) >droPer1.super_73 93353 80 + 267513 UCAUGGAUGAAAAAAAAGAGUUAUUAACGGAAGUACAUUCACCUGAUCAGUUGAAUGAACAUGCGCCUCUCCACACUGUU ...((((.((................((....)).((((((.((....)).))))))..........))))))....... ( -13.70, z-score = -0.17, R) >droWil1.scaffold_180699 818662 51 - 2593675 ---------------------UAUUAACGGAAGUACAUUCACCUGAUCAGUUGAAUGAACAUGAGCUACGUU-------- ---------------------.....((....)).((((((.((....)).))))))...............-------- ( -9.10, z-score = -1.09, R) >droMoj3.scaffold_6496 1301867 65 + 26866924 ------CCGUAAAUAAAGAGUUAUUAACGGAAGUACAUUCACCUGAUCAGUUGAAUGAACACGCGCAGCGU--------- ------((((.(((((....))))).))))..((.((((((.((....)).)))))).))((((...))))--------- ( -16.50, z-score = -2.75, R) >droGri2.scaffold_15245 17882547 66 + 18325388 -----UCAUGCAUUAA-AAGUUAUUAACGGAAGUACAUUCACCUGAUCAGUUGAAUGAACACGCGCUGUGCA-------- -----...(((((...-..((.....((....)).((((((.((....)).))))))...)).....)))))-------- ( -14.00, z-score = -0.84, R) >droVir3.scaffold_12875 4512169 55 - 20611582 ----------------AAAGUUAUUAACGGAAGUACAUUCACCUGAUCAGUUGAAUGAACACGCGCCGCGC--------- ----------------...(((....((....)).((((((.((....)).))))))))).(((...))).--------- ( -11.10, z-score = -0.76, R) >consensus _____UCGUGCCUUAAAAAAUUAUUAACGGAAGUACAUUCACCUGAUCAGUUGAAUGAACAUGAGCUCUGCU________ ..........................((....)).((((((.((....)).))))))....................... ( -9.10 = -9.10 + -0.00)

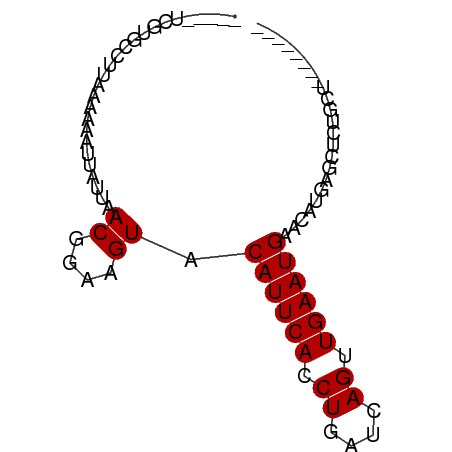
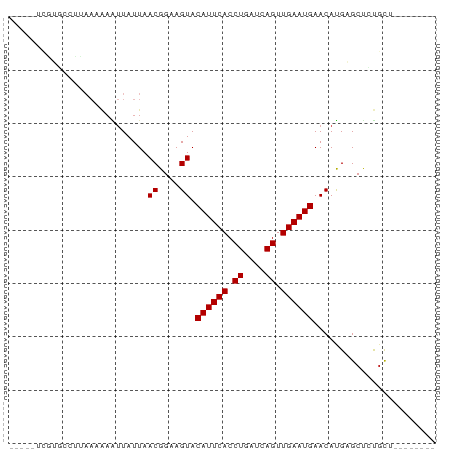
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:25:59 2011